Abstract
By their metabolic activities, microorganisms have a crucial role in the biogeochemical cycles of elements. The complete understanding of these processes requires, however, the deciphering of both the structure and the function, including synecologic interactions, of microbial communities. Using a metagenomic approach, we demonstrated here that an acid mine drainage highly contaminated with arsenic is dominated by seven bacterial strains whose genomes were reconstructed. Five of them represent yet uncultivated bacteria and include two strains belonging to a novel bacterial phylum present in some similar ecosystems, and which was named ‘Candidatus Fodinabacter communificans.’ Metaproteomic data unravelled several microbial capabilities expressed in situ, such as iron, sulfur and arsenic oxidation that are key mechanisms in biomineralization, or organic nutrient, amino acid and vitamin metabolism involved in synthrophic associations. A statistical analysis of genomic and proteomic data and reverse transcriptase–PCR experiments allowed us to build an integrated model of the metabolic interactions that may be of prime importance in the natural attenuation of such anthropized ecosystems.
Similar content being viewed by others
Introduction
Microorganisms are involved in biogeochemical nutrient cycles and have, therefore, a crucial role in the biosphere (Haferburg and Kothe, 2007; Falkowski et al., 2008; Konopka, 2009). Although microbial ecosystems constitute a major reservoir of our planet's genetic biodiversity, it is generally recognized that most of the microorganisms present in the environment are not accessible by the current culture-dependent techniques. The recent developments in genomics have given an unprecedented opportunity to gain insight into the structure and the functioning of microbial communities (Allen and Banfield, 2005; Bertin et al., 2008; Wilmes et al., 2009). Indeed, environmental genomics has extended their analysis far beyond the sole taxonomic studies, allowing the characterization of the whole community gene pool and its expression. Such approaches give rise to an integrated picture of ecosystems and are, therefore, of great interest to interpret the metabolisms present in ecological niches considered as extreme, including those impacted by human activities. Among the most toxic anthropogenic contaminants, arsenic is at the origin of serious forms of water pollution in industrial and post-industrial areas all over the world. In particular, high arsenic contents are encountered in mine drainage waters, where the metalloid is usually associated with sulfur, iron and other metals (Vaughan, 2006). Few forms of life are known to thrive in these often acid waters, as compared with neutral waters (Johnson and Hallberg, 2003; Denef et al., 2010). In this respect, the As(III) form (arsenite) can provide chemolithotrophic organisms with energy (Silver and Phung, 2005; Stolz et al., 2006).
The former mine of Carnoulès, Gard (France) provides an outstanding example of such an extreme environment. The sulfurous wastes contain As-rich pyrite and the leached waters are the source of a small stream called the Reigous that contains between 50 and 350 mg l−1 of soluble arsenic, mainly in the form of As(III) (Casiot et al., 2003; Egal et al., 2010). However, although the arsenic levels remain still high, this concentration decreases by 95% between the source of the Reigous and its confluence with the river Amous, 1.5 km downstream. This natural process of attenuation seems to be mainly due to microbial metabolism, leading to the oxidation of iron and arsenic into Fe(III) and As(V), and their co-precipitation with sulfur. In addition, laboratory experiments suggest that bacteria belonging to Thiomonas or Acidithiobacillus genera are involved in this process in situ (Bruneel et al., 2003; Casiot et al., 2003; Duquesne et al., 2003; Morin et al., 2003; Egal et al., 2009). However, 16S-based community analyses have revealed that other genera are present in this ecosystem (Duquesne et al., 2003, 2008; Bruneel et al., 2003, 2006, 2008, 2011). The biological activity of these uncultured bacteria may have a significant impact on the functioning of this ecosystem. In the present study, we used a multidisciplinary approach that took advantage of the ‘omics’ methods, to decipher the role of microorganisms, including uncultured bacteria, in the complex metabolic processes at work in the Carnoulès acid mine drainage (AMD), an arsenic-rich ecosystem.
Materials and methods
Sampling and chemical analysis
Samples were collected in May 2007 at the station called COWG (Carnoulès Oxydizing Wetland, G) located 30 m downstream of the spring (Bruneel et al., 2003). In all, 5 cm deep white sediments were collected at the bottom of the creek using a sterile tube and pooled (Global positioning system (GPS) coordinates: 44°07′01.80″N/4°00′06.90″E), while the running water (that is a thin column, <10 cm) covering these sediments was collected in triplicate and filtered (300 ml) through sterile 0.22 μm nuclepore filters. These filters were transferred into a collection tube, frozen in liquid nitrogen and stored at −80 °C until further analysis. The main physico-chemical parameters (pH, T°, dissolved oxygen) were determined in the field and arsenic speciation, Fe(II) and sulfate analyses were performed as previously described (Bruneel et al., 2008).
DNA isolation and sequencing
DNA was extracted from the cellular fraction either directly using the UltraClean Soil DNA Isolation kit (MoBio Laboratories Inc., Carlsbad, CA, USA), or after separation of microbial cells by Nycodenz gradient, using the Wizard Genomic DNA Extraction kit (Promega, Madison, WI, USA) and stored at −20 °C (Supplementary Information). Nebulized DNA fragments ranging from 3 to 5 kb were used to construct a genomic library and DNA inserts were sequenced, as previously described (Muller et al., 2007), giving rise to 550 920 Sanger reads. In parallel, 281 758 DNA reads were obtained by GS-FLEX pyrosequencing using standard procedures. Both methods produced a total of 430.3 Mbp.
Clone library and phylogenetic analyses
Bacterial diversity was analyzed by cloning and sequencing PCR-amplified 16S rRNA genes (Supplementary Information). Sequences were compared with the RDP database (http://rdp.cme.msu.edu) (Wang et al., 2007) and BLAST (Basic Local Alignment Search Tool) online searches (Altschul et al., 1997). Phylogenies were constructed with the molecular evolutionary genetics analysis v4.0 program (Tamura et al., 2007) using maximum composite-likelihood model and neighbor-joining algorithm. The sequences of clones CG determined in this study were submitted to the EMBL database and were given accession numbers FN391809 to FN391849.
Bioinformatics, statistical analysis and phylogenomic approach
Two-third of the metagenomic sequences were successfully organized in seven bins (Supplementary Information) and were then integrated into the MicroScope platform for the prediction of coding sequences, followed by automatic and expert annotation (Vallenet et al., 2009). The mean polymorphism frequency in the population was assessed using SNIPer (Ning et al., 2001). Molecular phylogenies were inferred using 27 universal marker genes chosen from a reference gene set (Ciccarelli et al., 2006) (Supplementary Information). For each marker, the corresponding family of homologous genes from the HOGENOM database (Perrière et al., 2000) was identified. Each family data set was then aligned with the program MUSCLE (Edgar, 2004) and filtered using the program GBLOCKS (Castresana, 2000; Talavera and Castresana, 2007). Maximum-likelihood phylogenies were reconstructed with PhyML (v2.4.4; Guindon and Gascuel, 2003) using the Jones–Taylor–Thornton model of amino acid substitution (Jones et al., 1992). The metabolic network was predicted by the ‘Pathway Tools’ software (Karp et al., 2002) using MetaCyc (Caspi et al., 2008) as a reference pathway database and then analyzed by a multiple factor analysis from a two-dimensional matrix (Supplementary Information). DNA sequences were submitted to GenBank and were given the following ProjectIDs: 38045—Carnoules metagenome study (top level); 41535—Carnoules metagenome study, bin 1; 41537—Carnoules metagenome study, bin 2; 41539—Carnoules metagenome study, bin 3; 41541—Carnoules metagenome study, bin 4; 41543—Carnoules metagenome study, bin 5; 41545—Carnoules metagenome study, bin 6; 41549—Carnoules metagenome study, bin 7.
Protein extraction, gel electrophoresis and mass spectrometry identification
Proteins were extracted from cells recovered using Nycodenz gradient (Supplementary Information), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% gradient slab gels (PROTEAN II, Bio-Rad Laboratories, Hercules, CA, USA), stained with Coomassie Brilliant Blue R-250 and in gel digested, as previously described (Weiss et al., 2009). The resulting peptide extracts were analyzed by liquid chromatography and mass spectrometry (nanoLC-MS/MS) on a nanoACQUITY Ultra-Performance-LC (UPLC, Waters, Milford, MA, USA) coupled to SYNAPT hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Waters). Data were analyzed using the MASCOT 2.2.0. algorithm (Matrix Science, London, UK) to search against a target-decoy protein database (Supplementary Information).
PCR and reverse transcription
Total RNA was extracted as described (González-Mendoza et al., 2009) after Nycodenz gradient density separation of the bacterial community for 15 min at 4 °C from sediments stored at −80 °C. Total RNA was purified using the RNEasy Mini kit (Qiagen, Valencia, CA, USA) and digested with DNAse (Fermentas, Glen Burnie, MD, USA). The reverse transcription of punA, soxC and cobS transcripts was performed using SuperScript III One-Step reverse transcriptase–PCR System (Invitrogen, Carlsbad, CA, USA) and 5′-CTCCATCCGAAAAAGTGCTC-3′/5′-AAAGAGGTTTTGCTGCGGTA-3′, 5′-ATCGACGGATTTCTGGATTG-3′/5′-GGTAACGCTGCCATCTAAGC-3′ and 5′-TTGCTGATTTGTTGCTCTGG-3′/5′-TCCAGCAGACTGGACAACAC-3′ primers, respectively. A negative control was carried out without the first reverse transcription step. Amplified fragments were sequenced by Millgen and the resulting sequences were analyzed using BLAST program (Altschul et al., 1997).
Bacterial culture and growth medium
Iron oxidation was tested with Acidithiobacillus ferrooxidans B5 strain isolated from Carnoulès (Casiot et al., 2003). A total of, 5 × 106 cells ml−1 were grown at 30 °C for 5 days in 100:10 liquid medium (Schrader and Holmes, 1988) supplemented or not with 1 mg l−1 cobalamin (Sigma, St Louis, MO, USA) and shaken at 140 r.p.m. Iron oxidation was followed by the appearance of an orange precipitate and cellular growth was estimated by colony-forming unit (CFU) counting.
Results
Phylogeny and genome reconstruction
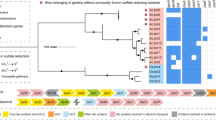
The main physico-chemical parameters (pH, T°, dissolved oxygen, iron concentration) were determined at the sampling site: a pH value of roughly 3.5 was measured, and arsenic, iron and sulfate concentrations reached 87, 625 and 3209 mg l−1, respectively, which correspond to 1.16 mM As, 11.16 mM Fe and 33.43 mM SO4 (Table 1). This further supports a persistent contamination by arsenic and other inorganic elements (Casiot et al., 2003; Egal et al., 2010). To identify the microbial species present in this ecosystem, DNA was extracted from the upper zone of sediments, 16S rRNA gene sequences were amplified, cloned and sequenced as described (Bruneel et al., 2008). Sequence analyses revealed a dominance of bacteria belonging to the Proteobacteria phylum (β- and γ-Proteobacteria with 38% and 23%, respectively) (Figure 1). Remaining clones belonged to Firmicutes (4%) and Acidobacteria (3%), and to a lesser extent to classes such as Spirochaetes (1%) and α-Proteobacteria (1%). This demonstrates that the microbial community at Carnoulès includes a few other bacterial phyla in addition to those previously identified (Bruneel et al., 2006, 2011).
Phylogenetic tree representing the taxonomic affiliation of the Carnoulès community microorganisms. The 16S rRNA gene sequences were obtained from DNA sediments after PCR amplification (clones CG-X) or metagenomic sequencing (CARN bins, except for CARN4, see text). A total of 759 positions in the final data set were obtained after all positions containing gaps and missing data were eliminated. The distances were computed using the maximum composite-likelihood method and the tree was inferred using the neighbor-joining method. The scale bar corresponds to 0.05 substitutions per site. Percentages of 1000 bootstrap resamplings that supported the branching orders in each analysis are shown above or near the relevant nodes. Bootstrap values are shown for branches with >50% bootstrap support.
To decipher the possible role of microorganisms in the biotransformation of toxic elements present at the study site, total DNA extracted from sediments was next fully sequenced. These genomic data were assembled in seven major bins called CARN1–CARN7, using a combination of 16S rRNA gene sequences, guanine and cytosine content (GC%), mean coverage of the various contigs, mean polymorphism frequency and similarity to already sequenced genomes, including At. ferrooxidans and Thiomonas sp. 3As (http://www.genoscope.cns.fr/agc/microscope/carnoulescope). Even though we demonstrated the existence of other species (see above), the presence of numerous dominant organisms would probably not have allowed to construct such large bins. Their size ranged from 1.5 to >4.0 Mbp and their guanine and cytosine content ranged from 52% to 65% (Table 2). DNA sequencing led to a low coverage for some contigs, in particular those of CARN3, which suggests that the size of the corresponding genome is underestimated. Finally, although the sequence assembling from ecotypes cannot be excluded, each supercontig group was considered to correspond to the genome of one single major organism (Supplementary Figure S1).
We evaluated the origin of the seven strains by a phylogenetic analysis of 16S rRNA sequences obtained from both the clone library and metagenome sequencing (Figure 1), except for CARN4, whose missing rRNA sequence precluded its 16S-based classification. Nevertheless, at the extremity of a contig, this bin contained a 23S rRNA gene sequence, which was similar to that of CARN1. We also performed a phylogenomic analysis of the bins using 27 universal marker genes (individually or combined) sufficiently conserved to build a tree of life (Ciccarelli et al., 2006), except for CARN6 where those markers were not found. Precisely, the candidates CARN2, CARN3 and CARN5 were affiliated to the β-Proteobacterium Thiomonas genus, the Acidobacteria clade and to the At. ferrooxidans γ-Proteobacterium, respectively (Supplementary Table S1). These phylogenetic results were supported by an analysis of gene order conservation known to be correlated with evolutive distances (Huynen and Bork, 1998). Finally, classification using 16S rRNA gene and RDP classifier indicated with confidence that CARN6 and CARN7 belong to the β-Proteobacteria class. In addition, 16S rRNA gene BLAST searches against NCBI nr database showed that the closest cultivated microorganisms to these two bins belong to the Thiobacillus and Gallionella genera, respectively (Supplementary Table S1).
Remarkably, the candidates corresponding to the CARN1 and CARN4 bins showed an important phylogenetic relationship and may represent two subpopulations according to the polymorphism distribution along their genome (Supplementary Figure S2). RDP classifier indicated that those bins did not correspond to any known taxonomic phylum and, according to the metabolic properties identified in the present study (see below), this new genus was herein named ‘Candidatus Fodinabacter communificans’ (from fodina, mine and communificare, share), in accordance with the recommendations for incompletely characterized microorganisms (Murray and Stackebrandt, 1995).
Metabolic potentialities of and in situ expressed activities by the seven dominant bacteria
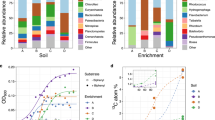
To determine the major metabolic potentialities of each bin, an in depth in silico analysis of their gene content was performed. In parallel, we investigated the functioning of the bacterial community by analyzing the proteins synthesized in situ by all bins. This metaproteomic approach allowed the reliable identification of >500 unique proteins belonging to various functional classes, for example membrane and transport, stress response and energy metabolism (Supplementary Tables S2ab). The experimental protein pattern was representative of the theoretical profile inferred from metagenomic data and the number of proteins identified from each bin was in agreement with the level of sequence coverage, which may reflect species abundance. Indeed, while a few proteins originated from CARN3 and CARN6, CARN1 and CARN4 bins were shown to express 70% of the identified proteins and emerged as abundant and very active in the ecosystem (Figure 2). Due to a low homology ranging from 20% to 30% with proteins present in databases, about 25% of the proteins synthesized by these last two bins were annotated as hypothetical (Supplementary Table S2b). One-third might, however, represent exported hydrolases, membrane transport proteins or sensors and modulators of chemotaxis and motility (data not shown).
Experimental metaproteomic pattern obtained by MS/MS identification of the proteins expressed in situ (CARN1, green; CARN2, orange; CARN3, black; CARN4, pink; CARN5, light blue; CARN6, brown and CARN7, blue). As a background, the theoretical distribution predicted from metagenomic data is represented in gray.
The genome of each bin was shown to contain at least one ars operon encoding arsenite efflux pumps and arsenate reductases. These genes are involved in arsenic resistance, and the presence of the corresponding proteins was demonstrated in protein extracts (Supplementary Table S2b). An arsM gene coding for an arsenite S-adenosylmethyltransferase was also identified in the genome of CARN6, in agreement with the presence of methylated forms of arsenic in Carnoulès sediments, that is monomethylarsonate and dimethylarsinate (7.10−4 and 3.10−4 mg, respectively, per mg dry weight). A few arsM homologous genes were also identified in unassembled sequences, which further supports the existence of arsenic methylation at the study site (data not shown). Finally, because of the structural similarity between As(V) and phosphate, arsenic metabolizing strains may preferentially transport phosphate via the specific Pst phosphate transport system rather than the Pit general transport mechanism, in order to reduce the entry of As(V). Accordingly, no Pit protein was identified in metaproteomic data, while several Pst proteins were detected (Supplementary Table S2b).
The microbial response to arsenic is known to result in various biological effects, including oxidative stress, DNA damage, exopolysaccharide synthesis and biofilm formation (Beyersmann and Hartwig, 2008; Marchal et al., 2010). Metaproteomic data showed that Carnoulès strains indeed express functions protecting against general and oxidative stress, including superoxide dismutase or thioredoxin, and chaperones such as DnaK or GroEL, which have previously been detected in AMD (Ram et al., 2005; Bruneel et al., 2011). Several proteins such as RecA and DPS involved in DNA recombination and repair were also identified. In addition, several flagellar proteins synthesized by CARN1 and CARN4 were identified in protein extracts (Supplementary Table S2b), as well as a protein involved in the synthesis of type 4 pilus expressed by Thiomonas sp. and known to have a role in twitching motility and adhesion (Li et al., 2010). The corresponding operon, found in the genome of this bin, was also present in the genomes of CARN5 and CARN7. These adaptive processes typically depend on multiple regulatory mechanisms that allow bacteria to respond to a wide panel of stimuli. This was supported by the proteomic identification of proteins such as the nucleoid-associated proteins HU and H-NS, which have a major role in both the structure and the function of chromosomal DNA (Tendeng and Bertin, 2003; Grove, 2010). Several regulators, including alternative σ factors such as σH and regulatory proteins belonging to the two-component systems or those involved in cell-to-cell signaling, were also expressed by various strains (Supplementary Tables S2ab), in agreement with the presence in their genome of multiple genes coding for proteins involved in stress response.
In a partly oligotrophic environment, such as the Carnoulès AMD, various metabolic reactions may be needed for autotrophic metabolism. In this respect, CARN5 and CARN7 may fix nitrogen by the expression of nif genes, which encode a nitrogenase (Dixon and Kahn, 2004), although this oxygen-sensitive enzyme was not detected in protein extracts. In addition, carbon fixation depends on proteins such as ribulose 1,5-biphosphate carboxylase/oxygenase (involved in Calvin cycle) (Badger and Bek, 2008), carboxysome structural proteins and carbon monoxide dehydrogenase (involved in acetyl-coenzyme A synthesis). These enzymes were identified in protein extracts, in agreement with the presence of the corresponding genes in Thiomonas sp. (CARN2), Acidithiobacillus sp. (CARN5) or Gallionella-related (CARN7) bins. These observations suggest that these three strains have a key role as organic compound primary producers. Nevertheless, other microorganisms may preferentially metabolize and recycle some organic compounds released by others. For example, the CARN6 bin carries the cellulase-encoding gene bczS and the α-amylase amyM gene, suggesting that this strain is able to metabolize complex carbohydrates. Similarly, several enzymes required for amino acid transport and metabolism were identified in the bins lacking the carbon and nitrogen fixation genetic determinants, in agreement with a mixotrophic or organotrophic metabolism. In particular, the CARN1 and CARN4 bins of ‘Candidatus Fodinabacter communificans’ contain genes such as liv and opp involved in branched amino acid and oligopeptide transport, respectively. In addition, both strains were shown to encode multiple peptidases, such as the metallopeptidase M61 and the serine protease S41 (Supplementary Table S2b), and to express the punA gene (Supplementary Figure S3), which suggests that they can use purine as a carbon source (Schuch et al., 1999). The CARN1 and CARN4 bins also expressed Gcv proteins of the glycine cleavage system that catalyzes the degradation of glycine and PdxS proteins involved in active vitamin B6 biosynthesis, which has an important role in amino acid metabolism (Fitzpatrick et al., 2007).
A wide diversity of bioenergetic electron chains may be needed to accommodate the presence in this ecosystem of various electron acceptors such as O2 or Fe(III) oxides and electron donors such as Fe(II) or sulfides. Accordingly, several terminal oxidases might be operative in all strains of the Carnoulès community, for example the cytochrome oxidase cox and cta operons and the cyo and cyd operons encoding quinol oxidases. Some operons involved in anaerobic respiration were also identified, in particular ntr, nar and nas in CARN2 and CARN7 bins, which suggests the possible recourse to anaerobic nitrate and nitrite metabolism. Nevertheless, the identification of several nicotinamide adenine dinucleotide-ubiquinone and cytochrome c oxidases in protein extracts supports the existence of an active aerobic metabolism at the sampling site (Supplementary Table S2b). The oxidation of H2 to protons may be a source of energy due to the presence in CARN2, CARN5 and CARN6 of several hydrogenase-encoding genes, in particular hox and hup operons (Friedrich and Schwartz, 1993). In addition, possible inorganic electron donors may comprise sulfur compounds. This is suggested by the presence and the expression (Supplementary Figure S3) in Thiomonas sp. (CARN2), Acidithiobacillus sp. (CARN5) or Gallionella-related sp. (CARN7) of genes such as hdr, sor, sox, tetH or sqr involved in the oxidation of reduced inorganic sulfur compounds, for example sulfide, sulfur, sulfite, thiosulfate or tetrathionate. Finally, the CARN7 bin harbors a dsrABEFHCMKLJOPN operon. The DsrA sequence showed only 39% identity with that of the sulfato-reducing Desulfovibrio vulgaris, but 72% with the amino acid sequence of the sulfo-oxidizing Thiobacillus denitrificans. This suggests that the Gallionella-related bin (CARN7) is able, like T. denitrificans, to use sulfur compounds as electron donors in its energy metabolism, as suggested in Gallionella ferruginea (Lutters-Czekalla, 1990).
Additionally, arsenite may constitute a possible inorganic electron donor and three arsenite oxidases with a nucleotide and amino acid sequence similarity >95% and 98%, respectively, and possibly involved in the bioenergetic transformation of As(III) to As(V) were shown to be expressed by Thiomonas sp. (CARN2). Although we cannot rule out that those sequences may originate from various ecotypes, the presence of multiple copies of aox operons has been recently demonstrated in Thiomonas strains (Arsène-Ploetze et al., 2010). Arsenate respiratory reductase-encoding gene, that is arrA, which allows anaerobic respiration of As(V), was neither identified in any of the seven bins nor in unassembled sequences, in agreement with the aerobic conditions prevailing in the ecosystem under study (Table 1). Importantly, the Acidithiobacillus sp. bin (CARN5) expressed the sole RusA protein (Supplementary Table S2b), which is involved in electron transport with iron used as an energy source. This further supports a major role of Acidithiobacillus sp. bin (CARN5) in the iron oxidation observed at Carnoulès (Bruneel et al., 2003; Duquesne et al., 2003; Morin et al., 2003). Interestingly, two cytochromes c synthesized by CARN7 were also identified in protein extracts (Supplementary Table S2b). They showed 61% and 43% amino acid similarity, respectively, with Cyc1 and Cyc2 of At. ferrooxidans. Cyc1 belongs to the c4 family and Cyc2 is an outer membrane cytochrome c proposed to receive electrons directly from ferrous iron (Yarzabal et al., 2002). Even though the other genes of the rus operon known to be involved in the electron transfer between Fe(II) and oxygen in At. ferrooxidans (Appia-Ayme et al., 1999) have not been detected in CARN7, it is tempting to hypothesize that this organism may be involved in iron cycling in the AMD under study.
Synecologic interactions within the microbial community
Autotrophic, mixotrophic or heterotrophic metabolisms present at Carnoulès suggest the existence of metabolic and nutrient exchanges that may be of prime importance inside the microbial community, revealing the importance of syntrophic relationships. To support and extend these observations, a multiple factor analysis of both metagenomic and metaproteomic data was performed on a two-dimensional matrix combining bins and enzymatic reactions. The most discriminant axes, that is capturing the highest dispersion in the resulting dot cloud, suggest an important and specific role for each organism inside the Carnoulès community. Indeed, factorial planes segregated CARN2 from the other bins (Figure 3a) and separated CARN1/CARN4 from CARN5/CARN7. In addition, CARN3 was opposed to CARN6, revealing CARN6-specific reactions (Figure 3b). No such correlation was observed between our data and those from the AMD biofilm (Tyson et al., 2004; Denef et al., 2010), which further supports the marked difference between these two ecosystems (Supplementary Figure S4). From the variable classification results, several clusters of reactions were then associated to each bin group (Figure 3). These include, for example, Cluster A4, which is linked to CARN2 (energy metabolism, inorganic nutrient metabolism and arsenic detoxification) (Supplementary Table S3a), and Cluster A1, which groups reactions common to CARN2 and CARN5 (Calvin–Benson–Bassham cycle and urea degradation pathways). In addition, Cluster A5 contains CARN6-specific reactions, such as those involved in cellulose metabolism, while Clusters A6, B1 and B4 include reactions related to lysine fermentation and to other amino acid or nucleoside degradation pathways in CARN1 and CARN4, and also gather reactions related to cobalamin biosynthesis (Supplementary Table S3b). Indeed, the CARN1 and CARN4 bins carry the cobSTU operon involved in cobalamin (vitamin B12) synthesis (Escalante-Semerena, 2007), which was shown by reverse transcriptase–PCR to be expressed in situ (Supplementary Figure S3). Remarkably, iron oxidation catalyzed by At. ferrooxidans was strongly increased in the presence of cobalamin (Supplementary Figure S5). These data suggest that this vitamin synthesized by CARN1 and CARN4 may be used by CARN5, leading to an increase in iron oxidation.
Multiple factorial analysis of the seven Carnoulès bins performed on a two-dimensional matrix combining bins and enzymatic reactions, respectively. To highlight possible metabolic distinctions between bins, three axes (F1 to F3) capturing the highest dispersion in the resulting dot cloud were selected; they represent more than half the total dispersion. Colored lines represent the projection of vectors corresponding to the enzymatic reaction frequencies, the external disk differentiating reactions whose enzymes were identified in the metaproteomic data. The reaction vectors were then hierarchically clustered, which led to 7 (A1 to A7) and 9 (B1 to B9) classes (indicated by the colors) for the first (a) and second (b) factorial planes, respectively. For each cluster, the corresponding reactions are listed in Supplementary Tables S3ab. For example, several reactions of Cluster A4 are concerned with energy and inorganic nutrient metabolism and in arsenic detoxification. As they are in the same quadrant of the plot as CARN2, these reactions are thus mostly specific to this bin. Similarly, Cluster A1 groups contain reactions common to CARN2 and CARN5 (Calvin–Benson–Bassham cycle and urea degradation pathways); Cluster A5 contains CARN6-specific reactions (cellulose metabolism) and Clusters A6, B1 and B4 include reactions in CARN1 and CARN4 (lysine fermentation other amino acid or nucleoside degradation pathways and cobalamin biosynthesis).
Discussion
In the last few years, a huge amount of genomic sequences have been published in databases, including a complete characterization of several bacteria metabolizing arsenic (Muller et al., 2007; Arsène-Ploetze et al., 2010). In microbial ecology, the major challenge remains, however, to determine more precisely the role of microorganisms and their relationships with other members of the microbial community that result in an efficient functioning of the ecosystem. In this respect, metagenomic approaches based on high-throughput technologies may be of great interest in synecology, since they allow to investigate the role of complex microbe consortia as a whole, including that of uncultured microorganisms in the processes taking place in situ. In the present study, we performed an in-depth analysis of descriptive and functional genomic data that allowed us to identify the dominant diversity present in an arsenic-contaminated ecosystem (Figure 4). Some microorganisms belong to bacterial genera already characterized, for example Thiomonas (CARN2) and Acidithiobacillus (CARN5). Others correspond, however, to uncultured microorganisms such as the one related to Gallionella sp. (CARN7) and the first representative of a novel bacterial phylum, here named ‘Candidatus Fodinabacter communificans’ (CARN1 and CARN4). Interestingly, only two 16S rRNA sequences (97% similarity) similar to CARN1 are present in databases and originate from other acid mine environments, suggesting that uncultivated bacteria of this genus are widespread in such ecosystems. Finally, the presence at the sampling site of two subpopulations of ‘Candidatus Fodinabacter communificans’ (Supplementary Figures S1 and 2) is reminiscent to what has been observed in other environmental contexts (Simmons et al., 2008; Wilmes et al., 2008). Such ecotypes, which may reflect an important genetic variability resulting from gene gain or loss, has been recently described in Thiomonas strains (Arsène-Ploetze et al., 2010).
Model of the Carnoulès bacterial community highlighting the major functions identified by metagenome sequencing or metaproteome characterization. These activities include carbon and nitrogen fixation, energy metabolism, flagellum and capsule biosynthesis, amino acid transport and degradation, detoxification and stress response, arsenic and iron metabolism. The possible interactions between these microorganisms or with other chemical or biological compounds present on the study site are indicated by arrows. CARN bins are numbered from 1 to 7.
Metagenomic and metaproteomic data emphasized the diversity of mechanisms that may be important in situ, allowing the seven dominant bacteria to adapt their metabolic activities to changes in environmental conditions (Figure 4). One example concerns the ability of microorganisms to live in organized surface communities called biofilms, where aggregated cells are embedded in an exopolymeric matrix. Such a lifestyle confers on them an higher resistance to various environmental stresses (Harrison et al., 2007) and favors the physical interactions between the cells (Davey and O’toole, 2000). In addition, flagellum biosynthesis and motility are often known to have a role in the first steps of biofilm formation (Soutourina and Bertin, 2003). Metagenomics revealed that most bins contain multiple flagellar and pili genes and metaproteomics allowed us to identify the corresponding proteins in CARN1/CARN4 and in CARN2, respectively (Supplementary Table S2b). While no flagellar operon was identified in Acidithiobacillus sp. (CARN5), as it may be observed in already sequenced related bacteria (http://www.genoscope.cns.fr/agc/microscope/carnoulescope), this bin contains several genes, such as galE and pgm involved in capsular biosynthesis in At. ferrooxidans (Barreto et al., 2005), suggesting a role of this strain in the formation of a biofilm.
More importantly, combined with multiple factorial analysis of the data, our observations demonstrated both the metabolic specificity and partnerships that may exist inside an arsenic-rich environment at the benefit of the microbial community as a whole. These processes include the fixation of inorganic carbon and nitrogen by several strains, in particular those belonging to the Thiomonas (CARN2), Acidithiobacillus (CARN5) and Gallionella-related (CARN7) genera. Indeed, these strains were shown to synthesize proteins such as the Rubisco, that is ribulose-1,5-biphosphate carboxylase oxygenase. This enzyme of the Calvin cycle catalyzes the first step of CO2 fixation. In addition, the genome of CARN5 and CARN7 also contains the nif genes, which may encode a nitrogenase. This further supports a key role of these organisms in nitrogen cycle inside the Carnoulès community. Their autotrophic metabolisms may be essential to reach an equilibrium between auto- and heterotrophy providing other partners with organic nutrients. In this respect, the presence of fucP and exuT genes coding for L-fucose and hexuronate transporters, respectively, in CARN3 may support, at least in part, the carbon requirements of this organism. In addition, CARN6 carries the cellulase-encoding gene bczS and the α-amylase amyM gene, suggesting its ability to metabolize more complex carbohydrates.
Remarkably, our results highlighted the role of several members of the Carnoulès community in the recycling of both mineral and organic resources, such as arsenic, iron, sulfur, urea, vitamins, nucleosides and amino acids (Figure 4). In this respect, the arsenite oxidase activity expressed by Thiomonas sp. (CARN2), associated with metabolisms such as iron oxidation and sulfur oxidation by Acidithiobacillus sp. (CARN5) and the Gallionella-related strain (CARN7), seems to be of prime importance in the co-precipitation of these inorganic elements, leading to the partial but efficient attenuation of this arsenic-contaminated ecosystem (Bruneel et al., 2003; Casiot et al., 2003; Duquesne et al., 2003; Egal et al., 2009, 2010). Other microorganisms, in particular ‘Candidatus Fodinabacter communificans,’ may recycle organic compounds released by others or provide them with essential cofactors, improving their biomineralization activities. First, CARN1 and CARN4 bins expressed genes involved in branched-chain amino acid and oligopeptide transport. Second, and unlike other Carnoulès bins, their genome carries drm and punA genes, suggesting that they can use purine as a carbon source (Schuch et al., 1999). Third, genes involved in amino acid fermentation were also identified in both bins, for example those of the lysine pathway, which converts lysine into butyrate, acetate and ammonia (Kreimeyer et al., 2007). Fourth, the CARN1 and CARN4 bins of ‘Candidatus Fodinabacter communificans’ carry the arginase-encoding gene rocF involved in urea biosynthesis, while a complete urease-encoding ure operon was present in Thiomonas sp. bin (CARN2). Fifth, genes involved in cobalamin biosynthesis, including the cobSTU operon, were identified in both CARN1 and CARN4 bins, while BtuC cobalamin transporters are possibly encoded by other bins, such as Thiomonas (CARN2) and Acidithiobacillus sp. (CARN5), as it may be observed in already sequenced related bacteria (http://www.genoscope.cns.fr/agc/microscope/carnoulescope). In Rhodopseudomonas palustris strain TIE-1, a CobS-like protein has been previously shown to be involved in iron oxidation, but the mechanism remains unclear (Jiao et al., 2005). Interestingly, we showed that cobalamin strongly activates iron oxidation in At. ferrooxidans, the first step in the natural remediation observed at the study site. Such a metabolic cooperation may thus be of great importance in the natural biomineralization observed at Carnoulès. Finally, cobalamin biosynthesis may also be useful to eukaryotes, in particular Euglena sp. present at the study site (Casiot et al., 2004) whose cell cycle is known to require this cofactor (Olaveson and Stokes, 1989; Bré et al., 1981).
Taken together, our data provide evidence that at least seven bacteria are involved in the functioning of the AMD ecosystem under study. In particular, our observations support the existence of multiple metabolic cooperations between the Carnoulès microorganisms (Figure 4). They also highlight an indirect but crucial role of the first representative of a novel and uncultured bacterial phylum, that is ‘Candidatus Fodinabacter communificans,’ in the biomineralization processes in this arsenic-rich ecosystem. In the future, descriptive and functional genomic approaches such as those presented here should give an integrated view of the biological objects present in any environment, their relationships, their role in the nutrient biogeochemical cycles and their possible use in the development of novel methods of bioremediation. More routinely used, such strategies will lead to important advances in microbial ecology, revealing what was recently regarded as impossible to explore.
References
Allen EE, Banfield JF . (2005). Community genomics in microbial ecology and evolution. Nat Rev Microbiol 3: 489–498.
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W et al. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Appia-Ayme C, Guiliani N, Ratouchniak J, Bonnefoy V . (1999). Characterization of an operon encoding two c-type cytochromes, an aa3-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl Environ Microbiol 65: 4781–4787.
Arsène-Ploetze F, Koechler S, Marchal M, Coppée J-Y, Chandler M, Bonnefoy V et al. (2010). Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet 6: e1000859.
Badger MR, Bek EJ . (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exper Bot 59: 1525–1541.
Barreto M, Jedlicki E, Holmes DS . (2005). Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl Environ Microbiol 71: 2902–2909.
Bertin PN, Médigue C, Normand P . (2008). Advances in environmental genomics: towards an integrated view of micro-organisms and ecosystems. Microbiology 154: 347–359.
Beyersmann D, Hartwig A . (2008). Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch Toxicol 82: 493–512.
Bré MH, El Ferjani E, Lefort-Tran M . (1981). Sequential protein-dependent steps in the cell cycle initiation and completion of division in vitamin B12 replenished Euglena gracilis. Protoplasma 108: 301–318.
Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personné J-C . (2006). Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoulès, France. Appl Environ Microbiol 72: 551–556.
Bruneel O, Pascault N, Egal M, Bancon-Montigny C, Goñi-Urriza MS, Elbaz-Poulichet F et al. (2008). Archaeal diversity in a Fe-As rich acid mine drainage at Carnoulès (France). Extremophiles 12: 563–571.
Bruneel O, Personné JC, Casiot C, Leblanc M, Elbaz-Poulichet F, Mahler BJ et al. (2003). Mediation of arsenic oxidation by Thiomonas sp. in acid-mine drainage (Carnoulès, France). J Appl Microbiol 95: 492–499.
Bruneel O, Volant A, Gallien S, Chaumande B, Casiot C, Carapito C et al. (2011). Characterization of the active bacterial community involved in natural attenuation processes in arsenic-rich creek sediments. Microb Ecol; e-pub ahead of print 12 February 2011.
Casiot C, Bruneel O, Personné JC, Leblanc M, Elbaz-Poulichet F . (2004). Arsenic oxidation and bioaccumulation by the acidophilic protozoan, Euglena mutabilis, in acid mine drainage (Carnoules, France). Sci Total Environ 320: 259–267.
Casiot C, Morin G, Juillot F, Bruneel O, Personné JC, Leblanc M et al. (2003). Bacterial immobilization and oxidation of arsenic in acid mine drainage (Carnoulès creek, France). Water Res 37: 2929–2936.
Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M et al. (2008). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 36: D623–D631.
Castresana J . (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552.
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P . (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287.
Davey ME, O’toole GA . (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867.
Denef VJ, Mueller RS, Banfield JF . (2010). AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4: 599–610.
Dixon R, Kahn D . (2004). Genetic regulation of biological nitrogen fixation. Na Rev Microbiol 2: 621–631.
Duquesne K, Lebrun S, Casiot C, Bruneel O, Personné JC, Leblanc M et al. (2003). Immobilization of arsenite and ferric iron by Acidithiobacillus ferrooxidans and its relevance in acid mine drainage. Appl Env Microbiol 69: 6165–6173.
Duquesne K, Lieutaud A, Ratouchniak J, Muller D, Lett MC, Bonnefoy V . (2008). Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ Microbiol 10: 228–237.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Egal M, Casiot C, Morin G, Elbaz-Poulichet F, Cordier MA, Bruneel O . (2010). An updated insight into the natural attenuation of As concentrations 3 in Reigous Creek (southern France). Appl Geochem 25: 1949–1957.
Egal M, Casiot C, Morin G, Parmentier M, Bruneel O, Lebrun S et al. (2009). Kinetic control on the formation of tooeleite, schwertmannite and jarosite by Acidithiobacillus ferrooxidans strains in an As(III)-rich acid mine water. Chem Geol 265: 432–441.
Escalante-Semerena JC . (2007). Conversion of cobinamide into adenosylcobamide in Bacteria and Archaea. J Bacteriol 189: 4555–4560.
Falkowski PG, Fenchel T, Delong EF . (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039.
Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T . (2007). Two independent routes of denovo vitamin B6 biosynthesis: not that different after all. Biochem J 407: 1–13.
Friedrich B, Schwartz E . (1993). Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Ann Rev Microbiol 47: 351–383.
González-Mendoza D, Morales-Trejo A, Brito-Vera H . (2009). A rapid and efficient method for isolation of total RNA from Euglena gracilis (Euglenoidea). Genet Mol Res 8: 482–486.
Grove A . (2010). Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol 13: 1–12.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Haferburg G, Kothe E . (2007). Microbes and metals: interactions in the environment. J Basic Microbiol 47: 453–467.
Harrison JJ, Ceri H, Turner RJ . (2007). Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microb 5: 928–938.
Huynen MA, Bork P . (1998). Measuring genome evolution. Proc Natl Acad Sci USA 95: 5849–5856.
Jiao Y, Kappler A, Croal LR, Newman DK . (2005). Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl Environ Microbiol 71: 4487–4496.
Johnson DB, Hallberg KB . (2003). The microbiology of acidic mine waters. Res Microbiol 154: 466–473.
Jones DT, Taylor WR, Thornton JM . (1992). The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282.
Karp P, Paley S, Romero P . (2002). The pathway tools software. Bioinformatics 18: S1–S8.
Konopka A . (2009). What is microbial community ecology? ISME J 3: 1223–1230.
Kreimeyer A, Perret A, Lechaplais C, Vallenet D, Médigue C, Salanoubat M et al. (2007). Identification of the last unknown genes in the fermentation pathway of lysine. J Biol Chem 282: 7191–7197.
Li Y-Q, Wan D-S, Huang S-S, Leng F-F, Yan L, Ni Y-Q et al. (2010). Type IV Pili of Acidithiobacillus ferrooxidans are necessary for sliding, twitching motility, and adherence. Curr Microbiol 60: 17–24.
Lutters-Czekalla S . (1990). Lithoautotrophic growth of the iron bacterium Gallionella ferruginea with thiosulfate or sulfide as energy source. Arch Microbiol 154: 417–421.
Marchal M, Briandet R, Koechler S, Kammerer B, Bertin PN . (2010). Effect of arsenite on swimming motility delays surface colonization in Herminiimonas arsenicoxydans. Microbiology 156: 2336–2342.
Morin G, Juillot F, Casiot C, Bruneel O, Personné JC, Elbaz-Poulichet F et al. (2003). Bacterial formation of tooeleite and mixed arsenic(III) or arsenic(V)-iron(III) gels in the Carnoulès acid mine drainage, France. A XANES, XRD, and SEM study. Environ Sci Technol 37: 1705–1712.
Muller D, Médigue C, Koechler S, Barbe V, Barakat M, Talla E et al. (2007). A tale of two oxidation states: bacterial colonization of arsenic-rich environment. PLoS Genet 3: e53.
Murray RGE, Stackebrandt E . (1995). Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45: 186–187.
Ning Z, Cox AJ, Mullikin JC . (2001). SSAHA: a fast search method for large DNA databases. Genome Res 11: 1725–1729.
Olaveson MM, Stokes PM . (1989). Responses of the acidophilic alga Euglena mutabilis (Euglenophyceae) to carbon enrichment at pH 3. J Phycol 25: 529–539.
Perrière G, Duret L, Gouy M . (2000). HOBACGEN: database system for comparative genomics in bacteria. Genome Res 10: 379–385.
Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC et al. (2005). Community proteomics of a natural microbial biofilm. Science 308: 1915–1920.
Schrader JA, Holmes DS . (1988). Phenotypic switching of Thiobacillus ferrooxidans. J Bacteriol 170: 3915–3923.
Schuch R, Garibian A, Saxild HH, Piggot PJ, Nygaard P . (1999). Nucleosides as a carbon source in Bacillus subtilis: characterization of the drm-pupG operon. Microbiology 145: 2957–2966.
Silver S, Phung LT . (2005). Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71: 599–608.
Simmons SL, DiBartolo G, Denef VJ, Goltsman DSA, Thelen MP, Banfield JF . (2008). Population genomic analysis of strain variation in Leptospirillum Group II bacteria involved in acid mine drainage formation. PLoS Biol 6: e177.
Soutourina OA, Bertin PN . (2003). Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27: 505–523.
Stolz JF, Basu P, Santini JM, Oremland RS . (2006). Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60: 107–130.
Talavera G, Castresana J . (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577.
Tamura K, Dudley J, Nei M, Kumar S . (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Tendeng C, Bertin PN . (2003). H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 11: 511–518.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A et al. (2009). MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009: bap021.
Vaughan DJ . (2006). Arsenic. Elements 2: 71–75.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Weiss S, Carapito C, Cleiss J, Koechler S, Turlin E, Coppee J-Y et al. (2009). Enhanced structural and functional genome elucidation of the arsenite-oxidizing strain Herminiimonas arsenicoxydans by proteomics data. Biochimie 91: 192–203.
Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B et al (2008). Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J 2: 853–864.
Wilmes P, Simmons SL, Denef VJ, Banfield JF . (2009). The dynamic genetic repertoire of microbial communities. FEMS Microbiol Rev 33: 109–132.
Yarzabal A, Brasseur G, Ratouchniak J, Lund K, Lemesle-Meunier D, DeMoss JA et al. (2002). The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J Bacteriol 184: 313–317.
Acknowledgements
AH-S, JC-A and DH were supported by a grant from the French Ministry of Education and Research and MM and FG-C by a grant from the Agence Nationale de la Recherche (ANR). Financial support came from the Université de Strasbourg (UdS), the Consortium National de Recherche en Génomique (CNRG), the Centre National de la Recherche Scientifique (CNRS) and the Agence Nationale de la Recherche (ANR) in the frame of the RARE and MicroScope projects. This work was done in the frame of the ‘Groupement de Recherche—Métabolisme de l’Arsenic chez les Microorganismes (GDR2909-CNRS)’ (http://gdr2909.alsace.cnrs.fr/).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Bertin, P., Heinrich-Salmeron, A., Pelletier, E. et al. Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteo-genomics. ISME J 5, 1735–1747 (2011). https://doi.org/10.1038/ismej.2011.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.51
Keywords
This article is cited by
-
Genome-resolved metagenomics inferred novel insights into the microbial community, metabolic pathways, and biomining potential of Malanjkhand acidic copper mine tailings
Environmental Science and Pollution Research (2023)
-
Alkaline extract from vermicompost reduced the stress promoted by As on maize plants and increase their phytoextraction capacity
Environmental Science and Pollution Research (2022)
-
Water and soil contaminated by arsenic: the use of microorganisms and plants in bioremediation
Environmental Science and Pollution Research (2022)
-
Synergistic Impacts of Arsenic and Antimony Co-contamination on Diazotrophic Communities
Microbial Ecology (2022)
-
Recent progress in the application of omics technologies in the study of bio-mining microorganisms from extreme environments
Microbial Cell Factories (2021)