Abstract
The bacterial symbiont Verminephrobacter eiseniae colonizes nephridia, the excretory organs, of the lumbricid earthworm Eisenia fetida. E. fetida transfers V. eisenia into the egg capsule albumin during capsule formation and V. eiseniae cells migrate into the earthworm nephridia during embryogenesis, where they bind and persist. In order to characterize the mechanistic basis of selective tissue colonization, methods for site-directed mutagenesis and colonization competence were developed and used to evaluate the consequences of individual gene disruptions. Using these newly developed tools, two distinct modes of bacterial motility were shown to be required for V. eiseniae colonization of nascent earthworm nephridia. Flagella and type IV pili mutants lacked motility in culture and were not able to colonize embryonic earthworms, indicating that both twitching and flagellar motility are required for entrance into the nephridia.
Similar content being viewed by others
Introduction
Animals and plants form cooperative, beneficial associations with bacteria that range from highly specific and obligate to diverse and facultative. Most animal–bacteria symbioses are initiated in early stages of life as juveniles acquire proper bacteria from either the environment or directly from the parent (Bright and Bulgheresi, 2010). This process presents challenges to a juvenile, as it must simultaneously attract appropriate bacteria while avoiding colonization by potential pathogens. For extracellular symbionts, this process of attracting, collecting and keeping the proper symbiont has been well described in very few systems established for laboratory study including the squid–Vibrio, leech–Aeromonas and nematode–Xenorhabdus associations (Nyholm and Mcfall-Ngai, 2004; Graf, 2005; Goodrich-Blair, 2007). These models are enabled by molecular tools to examine mechanisms governing these stages. Others have elegantly established developmental sequences for early colonization through observations in the environment or lab in the absence of molecular genetic tools (Nussbaumer et al., 2006; Davidson and Stahl, 2008). Without abilities to manipulate and test bacterial genes associated with early colonization processes, mechanisms for this critical phase of microorganism–host partnership remain vague.
Earthworms in the Lumbricidae harbor bacterial symbionts belonging to the Verminephrobacter genus in their nephridia, kidney-like osmoregulatory organs (Knop, 1926; Pinel et al., 2008; Lund et al., 2010). Distinct from the gut, nephridia are attached two per segment on either side of the gut. Each nephridium has a ciliated intake (nephrostome) leading to tubules forming three distinct regions. Fluid flows from the coelom through the nephrostome into a set of fine ciliated tubules leading into a second portion with a widened pouch, the ampulla, then to the bladder, and finally through a pore to the exterior (Figure 1a). Bacteria selectively colonize the ampulla. In Eisenia fetida, a widely distributed composting worm, three symbionts are known to colonize nephridia, Verminephrobacter eiseniae, a Flexibacter sp. and a Microbacteriaceae member (Davidson et al., 2010). Of these, only V. eiseniae can now be routinely maintained in culture independent of the host (Pinel et al., 2008). There is strong evidence for evolutionary longevity of the Verminephrobacter–earthworm association (Lund et al., 2010), including a process of vertical transmission, direct passage of symbionts from parent to offspring (Davidson and Stahl, 2006).
Anatomy of an earthworm nephridium. (a) Mature nephridium: 1, first loop, arrow heads indicate fluid flow through ciliated tubules; 2, second loop with ampulla containing bacteria; 3, bladder section before exit to exterior. (b) Colonization of nascent nephridium: 1, bacteria accumulate at a pore; 2, cells migrate into the colonization duct; 3, bacteria enter nephridia through a pore into the bladder then ampulla. a, ampulla; b, bladder; d; duct; n, nephrostome; p, pore.
During earthworm mating, bacteria are deposited into the nascent egg capsule. Embryos develop in a dense mixed population of bacteria and during embryogenesis, bacteria within egg capsules begin to colonize segments as they mature anterior to posterior (Davidson and Stahl, 2008). The capsule albumen of E. fetida contains specific symbiotic strains as well as other bacteria presumably entrained from soil (Davidson et al., 2010). Embryos are successfully colonized by specific bacteria from this mix and remain uncolonized by other resident bacteria. Prior microscopy studies have shown that bacterial cells initially accumulate near pores during the development of each segment. They then migrate into ducts that run dorsal to ventral on the lateral sides of each segment. Bacteria reside in ducts for a day or so, then complete migration by traveling into nascent nephridia, accumulating rapidly in the ampulla (Figure 1b). Only specific symbiotic bacteria enter the colonization duct, suggesting either specific attraction to the site or exclusion of incorrect cells (Davidson and Stahl, 2008). These observations suggest directed taxis of motile bacteria through albumen to the pore and into nascent nephridia. Although the physical path of colonization has been well visualized, the molecular mechanisms that facilitate colonization of earthworm embryos remain unknown.
Bacterial movement is essential for initiation of colonization for pathogens and beneficial bacteria in both plants and animals. The majority of well-studied examples are represented by pathogens, and includes the requirement of functional flagella and/or type IV pili (TFP) for pathogen colonization of the animal gut, human lung and plant leaf-surface (Haefele and Lindow, 1987; Nachamkin et al., 1993; Wassenaar et al., 1993). Beneficial bacterial colonization is less well known and only a few model systems have clearly demonstrated flagellar swimming motility to be essential for initiation of the association (Ames and Bergman, 1981; Malek, 1992; Graf et al., 1994). TFP provide adhesion and movement through extension, binding and then retraction of pili. Binding can be charge-dependent, or mediated by highly specific adhesins for binding to select receptors (Mattick, 2002). There are a few examples demonstrating TFP importance for symbiont colonization, but literature is sparse (Stabb and Ruby, 2003; Bohm et al., 2007).
Observations of the colonization process and evidence for motility of V. eiseniae in culture have led to the hypothesis that both flagella and TFP are involved during colonization of the earthworm embryo. The genome sequence of V. eiseniae EF01-2 contains all genes necessary for twitching and flagellar motility guided by chemotaxis (Pinel, 2009). Under the microscope, cells were observed spinning as if tethered on a glass slide, and single polar flagella were evident by transmission electron microscope (Pinel et al., 2008). In this paper, we describe application of genetic methods for study of mechanisms used by V. eiseniae to colonize embryos of the earthworm E. fetida. Specifically, we investigated the necessity of two forms of bacterial motility, flagellar swimming and TFP-mediated twitching, for successful colonization of the host.
Materials and methods
Bacterial strains and culture conditions
Although a genome sequence is available for V. eiseniae strain EF01-2, the strain EF05-2r (99.9% 16S rRNA sequence identity to EF01-2) was used for genetic studies because it lacks the extrachromosomal plasmid (pVEIS01), has a shorter generation time (Pinel et al., 2008) and a spontaneous rifampicin-resistant variant was available (EF05-2r). V. eiseniae EF05-2r was maintained on Acidovorax complex media (ACM) at 28 °C (Pinel et al., 2008). Escherichia coli TOP10 (Invitrogen, Carlsbad, CA, USA) and S17-1 (Simon et al., 1986) were maintained on Luria–Bertani agar at 37 °C. Kanamycin 30 or 50 μg ml−1, rifampicin 100 μg ml−1, and tetracycline 15 μg ml−1 were used as appropriate.
Mutant construction
The flgK gene encodes a flagellar hook-associated protein that forms the junction between the hook (FlgE) and filament monomers (FliC) (Berg, 2003). The neomycin phosphate transferase gene, npt2, conferring kanamyacin resistance was inserted into the flgK gene of V. eiseniae EF05-2r. Primers for amplification of 1.3-kb internal portion of the flgK gene were designed based on Veis_0560 (gb/CP000542.1/:628810-635405; flgKfrag-F-AVR2: 5′-GCCCTAGGCTGCAAACGACCGGCCACAACATTGCCA-3′; flgKfrag-R-HIND3: 5′-GCAAGCTTGGCCGCCATGACCGGGTTGGC-3′, underlined sequence denotes added restriction-enzyme cut sites) and used to amplify a portion of flgK from V. eiseniae EF05-2r genomic DNA. The fragment was cloned into a modified Gateway (Invitrogen) vector pENTR/D-Topo-MCS:kan (Shepherd and Lindow, 2009) as an AvrII/HindIII fragment. The resulting construct containing the flgK sequence and npt2 was transferred into pLVC-D suicide vector (Marco et al., 2005) via LR Clonase reaction (Invitrogen) to generate pLVC-DflgK, then introduced into donor E. coli strain S17-1 by electroporation (Simon et al., 1986). For biparental mating, S17-1 (pLVC-DflgK) and EF05-2r were mixed at a 1:10 ratio, plated on ACM and incubated overnight at 28 °C. Mating mixtures were harvested and plated on ACM containing kanamycin (30 μg ml−1) and rifampicin (100 μg ml−1). After 2–3 weeks of incubation at 28 °C, single colonies were restreaked for purity and tested for marker insertion.
In addition, a double crossover insertion was generated to remove coding regions of both the flgK and flgL genes in EF05-2r. Primers for amplification of 1-kb internal portion of the flgL gene were designed based on Veis_0559 (gb/CP000542.1/:628810-635405; flgLFow: 5′-CTCGAGCAGAACTTTCGCGCATTGGTGG-3′; flgLRev: 5′-TCTAGACAGCGACAATCTTTGCACCTGG-3′) and used to amplify a portion of flgL from V. eiseniae EF05-2r genomic DNA. The fragment was cloned into pENTR/D-Topo-MCS:kanflgK as a XhoI/XbaI fragment. The resulting construct containing npt2 flanked by flgK and flgL sequences was transferred into pLVC-D as previously described. pLVC-DflgKL was introduced into donor E. coli strain S17-1 by electroporation. Biparental mating and selection of mutants were done as previously described.
PilB and PilC are involved in export and assembly of TFP pilin monomers (Watson et al., 1996). Removal of either one or both results in loss of pilus construction (Nunn et al., 1990; Strom and Lory, 1993). Disruption of pilB (gb/CP000542.1/:4306572-4311008, Veis_3919) and pilC (Veis_3920) of V. eiseniae EF05-2r was generated by insertion of the kanamyacin-resistant marker. DNA fragments of ∼1 Kb in size were amplified from EF05-2r genomic DNA, using primers pilBfrag-F (5′-CCTAGGATTTACAAGAAGTCCCAGGCCAACCGCA-3′), pilBfrag-R (5′-AAGCTTTACCTGATTCACGCCCGGCAGGTTGATT-3′), pilCfrag-F (5′-CTCGAGAAAGTCTTCGAATGGGAAGGCAAGGACC-3′) and pilCfrag-R (5′-TCTAGACTTCACCAATGGCACACATTTGCAGCACCA-3′). The pilB sequence was cloned upstream of npt2 in pENTR/D-Topo-MCS:kan as a AvrII/HindIII fragment and the pilC sequence was subsequently cloned downstream of npt2 as a XhoI/XbaI fragment. The resulting construct containing npt2 flanked by pilB and pilC fragments was cloned into pLVC-D to create pLVC-DpilBC. Conjugation of EF05-2r was performed as above and mutants were screened for marker insertion. A mini-Tn5 transposon conferring kanamycin resistance was introduced into V. eiseniae EF05-2r from E. coli S17-1 (pRL27) (Larsen et al., 2002) through biparental mating, at a 1:10 ratio, O/N at 28 °C and subsequent selection on ACM with appropriate antibiotics to generate RTn5.1 (Random Tn5 clone1) and RTn5.2. All resulting mutants (VEflgK−, VEflgKL− and VEpilBC−) screened for resistance marker were confirmed using PCR to contain the appropriate insertion.
DNA sequencing
Genomic DNA of V. eiseniae EF05-2r was collected with a DNAeasy tissue kit (Qiagen, Germantown, MD, USA). Automated DNA sequencing was preformed at the University of Washington High Throughput DNA Sequencing Facility via illumina sequencing. The DNA sequence was assembled and open reading frames were assigned with a combination of the Glimmer3 algorithm (Delcher et al., 2007), GeneSifter (Geospiza, Seattle, WA, USA) and BLAST (Altschul et al., 1990). Accession numbers of DNA sequences: pilBCDcoaE (JN900250), flgIJKL (JN900251).
Motility assays
Cells were grown on ACM plates, harvested and washed in 10 mM KPO4 buffer, then 107 cells were stab inoculated into 0.3% agar ACM plates, incubated overnight at 28 °C and examined for cell migration through agar. Twitching motility was observed by stab inoculation into the bottom of 1.5% phytagel ACM plates and examined for movement along the petri dish bottom-surface. Strains were also harvested from plates, suspended in ACM broth (OD600≈1.0) and observed in suspended static culture at room temperature for adherence behavior, aggregation and binding to the culture tubes.
Electron microscopy
Cells were grown on ACM plates, collected and suspended in 1.25% glutaraldehyde, 0.1 M sodium phosphate buffer, pH 7.3, overnight at 4 °C, washed in buffer, and post-fixed with 2% osmium tetroxide, in 0.05 M phosphate buffer 1.5 h, rinsed and stored in 0.1 M phosphate buffer, pH 7.3. Samples for transmission electron microscope were mounted on 150 mesh rhodium/copper grids, stained with uranyl acetate and lead citrate and examined using a JEM 1200EX II TEM (JEOL Ltd, Tokyo, Japan).
Elimination and reestablishment of V. eiseniae from earthworm egg capsules and nephridia
Adult and juvenile E. fetida were maintained as previously described (Davidson and Stahl, 2006; Davidson et al., 2010). Egg capsules were collected and maintained on moistened filter paper in petri dishes. Bacterial symbionts were eliminated by treatment of egg capsules with antibiotics as previously described (Davidson and Stahl, 2006). In addition to 150 μg ml−1 kanamycin for elimination of V. eiseniae, 150 μg ml−1 erythromycin was used to eliminate both V. eiseniae and Flexibacter species. Hatchlings from each treatment were assayed for nephridial bacteria by fluorescence in situ hybridization (FISH; see next section) (n=10–15). Cured worms were maintained until sexual maturity and offspring monitored for nephridial bacteria. Capsules from cured adults were used to test colonization competence of mutants.
Cured capsules were inoculated with V. eiseniae EF05-2r by injection to reestablish nephridial bacteria. Eggs were first surface sterilized with 50% ethanol for 15 s, rinsed with sterile water three times and then allowed to dry until a small dimple appeared in the shell to accommodate additional fluid. Bacterial suspensions, approximately 1–3 μls, were injected into capsules with a 50 g needle and capsules were maintained in petri dishes at room temperature (∼20 °C). Eggs were partially submerged in diH20 to maintain humidity and hydration of eggs, and 30 μg ml−1 kanamycin was supplemented in suspension water to maintain selective pressure on mutants. At least 10 capsules were used per treatment, and at least 6 worms assayed from each treatment by FISH 2 days after hatching. Colonization of hatchlings was observed from 10 segments, each typically containing 2 nephridia, in the anterior, middle and posterior areas of hatchlings.
Optimal bacterial cell number needed to achieve full colonization was established by injecting a series of V. eiseniae EF05-2r cell solutions, ranging from 8 × 103 to 3.6 × 108 cells (∼0.002–100-fold average normal number), into capsules (0–1-day-old). The optimal time for injection for full colonization was tested by injecting the effective number of cells into capsules containing embryos staged at 0–3 days, 5–7 days and 10+ days of development. Cured capsules lacking both V. eiseniae and Flexibacter species or with Flexibacter sp. present were inoculated to test interference by precolonization of the nephridia by Flexibacter sp.
FISH for detection of nephridial colonization
Fixation and FISH were performed on intact hatchlings as described previously using stringent conditions established for each probe (Schramm et al., 2003; Davidson and Stahl, 2008). The following probes were used: LSB 145—V. eiseniae (Schweitzer et al., 2001; Schramm et al., 2003), EUB 338—bacterial domain (Amann, 1995) and Flexi 145—genus Flexibacter (Davidson et al., 2010). Specimens were mounted in Vectashield (Vector Labs, Inc., Burlingame, CA, USA) and fluorescence detected using excitation at the appropriate wavelength with a Zeiss LSM Pascal laser scanning confocal microscope (Carl Zeiss, Jena, Germany).
Results
Motility and aggregation of V. eiseniae
Flagellar motility
V. eiseniae possesses genes needed for flagella production. The gene cluster flgIJKL is arranged in an apparent operon in V. eiseniae EF01-2. Sequence analysis of V. eiseniae EF05-2r flgIJKL showed identical synteny and 99% sequence similarity. FlgK and FlgL function as flagellar hook-associated proteins that form the junction between the hook (FlgE) and filament monomers (FliC) (Berg, 2003). After 24 h of incubation, a band of V. eiseniae EF05-2r cells had clearly migrated away from the point of inoculation in soft agar plates (Figure 2a). Interruption of the flgKL coding sequences of EF05-2r was confirmed by PCR, after transformation via conjugation. The flagella mutants, VEflgK− (Figure 2a) and VEflgKL−(data not shown), failed to move away from the point of inoculation in semisolid media.
Motility tests of V. eiseniae wild-type and mutants. (a) Soft agar plate inoculated with V. eiseniae EF05-2r (left) and VEflgK− (right); (b) Phytagel plate stab inoculated with EF05-2r (left) and VEpilBC− (right). Arrowheads, point of inoculation; arrows, migrating front of cells. Note: label markings were digitally removed from A.
TFP-mediated motility
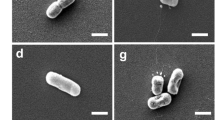
V. eiseniae also possesses genes required for TFP synthesis. The gene cluster pilBCDcoaE is arranged in an apparent operon in V. eiseniae EF01-2. Sequence analysis of V. eiseniae EF05-2r pilBCDcoaE showed identical synteny and 99% sequence similarity. The pilB and pilC genes encode components of the TFP machinery that export and polymerize PilA, pilin monomers. PilD cleaves PilA subunits prior to TFP assembly (Strom et al., 1993; Mattick, 2002). The function of coaE in TFP construction is undetermined. After stab inoculation, V. eiseniae EF05-2r cells were observed to move along the interface of the plastic bottom and the phytagel media (Figure 2b), a behavior characteristic of twitching motility. Transmission electron microscope analysis of cells from static cultures revealed rod-like structures extending as much as 4 μM away from the polar ends of wild-type (Figure 3a) VEflgK− and VEflgKL− cells moved between the plastic–phytagel interface and possessed rod-like structures identical to wild type (data not shown). Replacement of portions of coding regions of both pilB and pilC in EF05-2r, confirmed by PCR as above, eliminated twitching motility (Figure 2b) and cell surface pilus structures in VEpilBC− (Figure 3b).
Aggregation in liquid media
In static culture, the wild-type V. eiseniae EF05-2r cells aggregated and adhered weakly to the surface of plastic falcon tubes (Figure 4a). The flagellar mutants, VEflgK− (Figure 4b) and VEflgKL− (data not shown), adhered to the plastic surface as well. Although association with the tube wall was not stable, as gentle agitation easily displaced the aggregates, cell clumping was visible by light microscopy. Disruption of pilB and pilC resulted in loss of the aggregation phenotype, and cells failed to form clumps or bind to the sides of the tubes. VEpilBC− cells often remained dispersed in solution, and eventually settled to the bottom of the tube (Figure 4c).
Curing and reestablishment of V. eiseniae in earthworm nephridia
Nephridia of untreated hatchling worms are colonized by both V. eiseniae and Flexibacter sp. in the ampulla and only V. eiseniae in the bladder (Figure 5a). Treatments of capsules with 150 μg ml−1 kanamycin resulted in V. eiseniae-free hatchlings that retained Flexibacter sp. (Figure 5b). Treatment with 150 μg ml−1 erythromycin eliminated all nephridial symbionts, confirmed by FISH LSCM (Figure 5c) with EUB-338, Flexi 145 and LSB 145 probes, which resulted in no signal. Hatchlings from both treatments grew to sexual maturity.
FISH LSCM images of nephridia of E. fetida hatchlings. Hatchlings from (a) untreated capsule, (b) 150 μg ml−1 kanamycin treatment and (c) 150 μg ml−1 eurythromycin treatment. (d) Hatchlings from capsules lacking V. eiseniae inoculated with EF05-2r and (e) capsules lacking both symbionts inoculated with EF05-2r. (f) Hatchlings from V. eiseniae-free egg capsules inoculated with VEflgK− and (g) symbiont-cured egg capsules inoculated with VEflgK−. (h) Hatchlings from V. eiseniae-free egg capsules inoculated with VEpilBC− and (i) symbiont-cured egg capsules inoculated with VEpilBC−. Green, V. eiseniae; Red, Flexibacter sp. Scale bars, 50 μm; arrows, bladder; arrowheads, autofluorescent locomotive setae structures; circles, ampulla.
Capsules (0–1-day old), each containing approximately 5 μl of albumin, have an estimated 9x105 bacterial cells per μl of albumin, with approximately 3.2 × 106 V. eiseniae cells per capsule (Davidson and Stahl, 2008). In capsules lacking both V. eiseniae and Flexibacter sp., 8 × 105 and 8 × 106 EF05-2r cells (0.25 and 2.5 times the estimated normal population, respectively) were required for full colonization of hatchling nephridia (n=10 hatchlings, 60 nephridia per hatchling observed). Inconsistent colonization resulted from injection of 8 × 104 cells, and no colonization was observed with 8 × 103 EF05-2r cells. Embryos failed to develop following injection of cell numbers >100-fold the normal V. eiseniae population.
Injections of cultivated EF05-2r into egg capsules lacking both Flexibacter sp. and V. eiseniae resulted in full colonization regardless of the stage of embryos at time injection (n=6, 60 nephridia per E. fetida hatchling at each stage) (Table 1). In contrast, inoculations of EF05-2r into capsules containing Flexibacter sp. resulted in variable V. eiseniae colonization associated with developmental stage (Table 1). The excess (10 × ) number of injected V. eiseniae cells relative to naturally occurring numbers did not alter normal co-colonization of Flexibacter sp. symbiont regardless of embryonic stage at inoculation. Worms associated with only EF05-2r or EF05-2r and Flexibacter sp. were raised to sexual maturity and no developmental defects were observed in lab culture.
Assessment of mutant colonization competence
Capsules (0–3 days) inoculated with 5 × 107 EF05-2r cells yielded fully colonized hatchlings with V. eiseniae cells in the ampulla (circled) and bladder (arrows) of the nephridia, with either Flexibacter sp. present or absent (Figures 5d and e). Consistent with normal colonization, cells only transiently resided in the bladder. At maturity, bacteria were found only in the ampulla. Hatchlings from capsules injected with VEflgK− cells rarely showed Verminephrobacter cells in the ampulla and primarily only in the bladder, regardless of the presence or absence of Flexibacter sp. These cells failed to move beyond the bladder (Figures 5f and g). Juveniles examined 2–3 weeks post hatching did not contain VEflgK− cells in either bladder or ampulla. Flexibacter sp. colonization of the ampulla appeared normal. Hatchlings from capsules injected with VEflgKL− exhibited the same results.
The TFP mutant failed to colonize the earthworm nephridia regardless of the presence or absence of the Flexibacter sp. symbiont. Inoculation of 5 × 107 VEpilBC− cells into capsules (0–3 days) lacking V. eiseniae, or both V. eiseniae and Flexibacter sp., failed to show V. eiseniae cells in any region of the nephridia when examined by FISH LSCM (Figures 5h and i). Flexibacter sp. appeared in the ampulla as in controls. Repeated colonization experiments with identical, yet independently generated, mutations of the flagella and TFP mutants displayed the same colonization deficiencies (Table 2, n=6 hatchlings). Random Tn5 insertion mutants of V. eiseniae were used as controls to determine secondary effects of the npt2 insertion or presence of antibiotics throughout the colonization period. V. eiseniae strains RTn5.1 and RTn5.2 containing a random mini-Tn5 cassette displayed normal colonization in similar inoculations of egg capsules, appearing in both the ampulla and the bladder of hatchlings (Table 2, n=10 hatchlings).
Discussion
This work provides a significant advancement in development of a new model system for understanding mechanisms of non-pathogenic bacteria–host interactions. The results describe the first mechanisms identified for early stages of establishment of a symbiont in an embryonic host stage. Methods for targeted gene disruption were demonstrated for the first time for host species-specific symbiont V. eiseniae EF05-2r and used to show the requirement of two distinct mechanisms of motility for successful colonization of the host. Using the V. eiseniae EF01-2 genome to guide gene selection, both flagellar and TFP systems were successfully disrupted, as shown by the loss of flagellar motility with flgK and flgL mutations, and loss of twitching with pilB and pilC disrupted. These two modes of motility are necessary for V. eiseniae to complete the journey from albumin into the colonization duct, then into the bladder and finally the ampulla of nascent nephridia. Without flagella, but TFP intact, cells make it as far as the bladder, indicating that TFP are used to migrate into the duct, and bladder, but that flagella are required to make it the rest of the way. The failure of TFP mutants to enter the duct, even with flagellar motility intact, indicates that TFP are essential for successful migration into and/or through the colonization pore and duct (Figure 6). Use of redundant mutant strains producing identical phenotypes in culture and colonization experiments supports our conclusions and lessens the possibility of secondary mutations obfuscating colonization defects observed, despite the lack of a mutant complementation method.
These observations suggest two important features of this colonization process. First, surface features of the embryo and the colonization duct are critical for initial migration mediated by TFP-binding. These may be specific receptors that act to select bacteria with the proper adhesins. If chemotaxis is involved in directing the cells, it is by directed twitching motility. Second, swimming is required to get past the bladder and stay in the ampulla, suggesting an outward current produced by ciliated tubules in the nephridia is active at this stage and may serve as a barrier to bacterial cells that cannot swim. In addition to swimming, flagella can serve as an attachment point during colonization. Flagellar cap protein FliD of Pseudomonas aeurugenosa and Clostridium difficile has been shown to adhere to epitopes present in host tissues (Arora et al., 1998; Tasteyre et al., 2001), and flagellin monomer FliC of enteropathogenic and enterohemorrhagic E. coli has a strong binding affinity for mucin of host mucosal surfaces (Erdem et al., 2007). The polar orientation of V. eiseniae cells within the ampulla (Pinel et al., 2008) indicates possible binding interactions with polar flagella or TFP. If flagella are required to bind the ampulla wall, flagellar mutants would be cleared from the system and collected in the bladder before being eliminated.
The process of migration terminates with binding to the ampulla surface. The ampulla is not a blind end pouch, but tubules enter and exit the ampulla. The receptors here, or the chemical signals, lead to binding mediated by TFP, flagella and/or cell surface receptors, and a cessation of motility. Although both pili and fimbriae are among the better-characterized structures of microbial attachment (Mattick, 2002), fimbriae are not likely involved because neither fimbriae genes in the genome nor structures in micrographs were detected in Verminephrobacter. Pili are well described in pathogen–host interactions, including regulation of immune responses through binding to cell surface complement regulators, and persistence at colonization sites through binding (Kallstrom et al., 2001). However, there are few characterized beneficial bacteria TFP-mediated host interactions, but these examples support a critical role for TFP in early colonization. Similar to E. fetida (Davidson et al., 2010), hatchling squid acquire Vibrio spp. symbionts from a mixed microbial community, and pilA mutation in Vibrio fisheri generated a competitive disadvantage when challenged with wild type for colonization of light organ (Stabb and Ruby, 2003). For endophytic N2-fixing Azoarcus sp. BH72, TFP are necessary to initiate colonization of rice plants (Bohm et al., 2007). Although dense Verminephrobacter populations in the albumen (Davidson and Stahl, 2008) and movement of worm embryos throughout this suspension suggest the embryo surfaces would be in constant contact with bacteria, the TFP or flagella may enable the symbiont to maneuver through the albumen to the embryo surface and duct entrance. The nature of the binding specificity of V. eiseniae to surfaces of the earthworm embryo remains to be explored.
Important inter-specific bacterial interactions were noted during the development of this model system. Associations with other bacterial species have been documented in well-established model animal-symbiont systems. Aeromonas veronii associates with Rikenella-like bacteria in microcolonies that colonize the digestive tract of medicinal leech Hiurdo verbena (Kikuchi and Graf, 2007). V. fisheri outcompetes related nonsymbiotic species during colonization of squid hatchling mucus secretion in aggregates outside infection sites (Nyholm and McFall-Ngai, 2003). Although aggregates of Flexibacter sp. and V. eiseniae cells are observed in the albumen, associate at the entrance pore and are found in the colonization duct together (Davidson and Stahl, 2008; Davidson et al., 2010), observations from selective curing experiments indicate that V. eiseniae and Flexibacter sp. symbionts are not dependent on each other for colonization and persistence. Single member colonization does not rule out the possibility that both are working in concert in nephridia of mature worms under normal field conditions. Introduction of V. eiseniae later in development demonstrated that the presence of Flexibacter sp. can interfere with V. eiseniae colonization possibly by competitive exclusion. Flexibacter sp. has been visualized to be bound to ampulla cells as was V. eiseniae (Davidson et al., 2010). It is possible that Flexibacter sp. migration into nascent nephridia prior to V. eiseniae occupies binding sites too densely for co-colonization by late arrivals. Specific development events, such as change in host chemical cueing or duct architecture, may also limit colonization. Crypt cell morphology of the squid light organ is altered, increased microvillar density and cell swelling, post inoculation with Vibrio symbionts (Montgomery and McFall-Ngai, 1994; Lamarcq and McFall-Ngai, 1998), presumably increasing interactions to establish symbiosis. These observations indicate that synchronization of bacterial colonization in the embryo is important for full colonization. Preemptive colonization of biocontrol agent Pseudomonas fluorecens A506 reduces colonization of the Erwinia amylovera fire blight pathogen on pear nectaries (Wilson and Lindow, 1993). Previous observations showed that Flexibacter sp. may arrive earlier than V. eiseniae on occasion, but under normal egg capsule circumstances, V. eiseniae likely enters soon after to colonize the ampulla (Davidson et al., 2010).
Essential tools for analysis of mechanisms governing V. eiseniae interactions with the earthworm E. fetida during colonization were established in this study. This new model system adds to the limited number of symbiotic associations being studied that have one or both members amenable to genetic manipulation (Ruby, 2008). With this project, we have begun to study specific mechanisms of this association fulfilling some of molecular Koch's postulates (Falkow, 1988) for in-depth analysis of molecular mechanisms. This is a novel model based on molecular mechanisms of selective bacterial colonization during embryogenesis. Although much has been gleaned looking at the end point colonization of the nephridia in hatchling worms, further analysis throughout embryonic development is needed to refine mechanisms of the observed colonization deficiencies.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Amann RI . (1995). Fluorescently labeled, ribosomal-rna-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol 4: 543–553.
Ames P, Bergman K . (1981). Competitive advantage provided by bacterial motility in the formation of nodules by rhizobium-meliloti. J Bacteriol 148: 728–729.
Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R . (1998). The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun 66: 1000–1007.
Berg HC . (2003). The rotary motor of bacterial flagella. Ann Rev Biochem 72: 19–54.
Bohm M, Hurek T, Reinhold-Hurek B . (2007). Twitching motility is essential for endophytic rice colonization by the N-2-fixing endophyte Azoarcus sp strain BH72. Mol Plant Microbe Interact 20: 526–533.
Bright M, Bulgheresi S . (2010). A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8: 218–230.
Davidson SK, Stahl DA . (2006). Transmission of nephridial bacteria of the earthworm Eisenia fetida. Appl Environ Microbiol 72: 769–775.
Davidson SK, Stahl DA . (2008). Selective recruitment of bacteria during embryogenesis of an earthworm. ISME J 2: 510–518.
Davidson SK, Powell RJ, Stahl DA . (2010). Transmission of a bacterial consortium in Eisenia fetida egg capsules. Environ Microbiol 12: 2277–2288.
Delcher AL, Bratke KA, Powers EC, Salzberg SL . (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics (Oxford) 23: 673–679.
Erdem AL, Avelino F, Xicohtencatl-Cortes J, Giron JA . (2007). Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J Bacteriol 189: 7426–7435.
Falkow S . (1988). Molecular kochs postulates applied to microbial pathogenicity. Rev Infect Dis 10: S274–S276.
Goodrich-Blair H . (2007). They've got a ticket to ride: Xenorhabdus nematophila–Steinernema carpocapsae symbiosis. Curr Opin Microbiol 10: 225–230.
Graf J . (2005). Molecular requirements for the colonization of Hirudo medicinalis by Aeromonas veronii.. In: Overmann J (ed), Molecular mechanisms of symbiosis. Series: Progress in Molecular and Subcellular Biology. Springer-Verlag: Berlin.
Graf J, Dunlap PV, Ruby EG . (1994). Effect of transposon-induced motility mutations on colonization of the host light organ by vibrio-fischeri. J Bacteriol 176: 6986–6991.
Haefele DM, Lindow SE . (1987). Flagellar motility confers epiphytic fitness advantages upon pseudomonas-syringae. Appl Environ Microbiol 53: 2528–2533.
Kallstrom H, Gill DB, Albiger B, Liszewski MK, Atkinson JP, Jonsson AB . (2001). Attachment of Neisseria gonorrhoeae to the cellular pilus receptor CD46: identification of domains important for bacterial adherence. Cell Microbiol 3: 133–143.
Kikuchi Y, Graf J . (2007). Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl Environ Microbiol 73: 1984–1991.
Knop J . (1926). Bakterien und bakteroiden bei Oligochaeten. Z Morphol Ökol Tiere 6: 588–624.
Lamarcq LH, McFall-Ngai MJ . (1998). Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by vibrio fischeri. Infect Immun 66: 777–785.
Larsen RA, Wilson MM, Guss AM, Metcalf WW . (2002). Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178: 193–201.
Lund MB, Davidson SK, Holmstrup M, James S, Kjeldsen KU, Stahl DA et al. (2010). Diversity and host specificity of the Verminephrobacter-earthworm symbiosis. Environ Microbiol 12: 2142–2151.
Malek W . (1992). The role of motility in the efficiency of nodulation by rhizobium-meliloti. Arch Microbiol 158: 26–28.
Marco ML, Legac J, Lindow SE . (2005). Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ Microbiol 7: 1379–1391.
Mattick JS . (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56: 289–314.
Montgomery MK, McFall-Ngai M . (1994). Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development (Cambridge) 120: 1719–1729.
Nachamkin I, Yang XH, Stern NJ . (1993). Role of Campylobacter-Jejuni flagella as colonization factors for 3-day-old chicks—analysis with flagellar mutants. Appl Environ Microbiol 59: 1269–1273.
Nunn D, Bergman S, Lory S . (1990). Products of 3 accessory genes, Pilb, Pilc, and Pild, are required for biogenesis of pseudomonas-aeruginosa Pili. J Bacteriol 172: 2911–2919.
Nussbaumer AD, Fisher CR, Bright M . (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441: 345–348.
Nyholm SV, McFall-Ngai MJ . (2003). Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: The first site of symbiont specificity. Appl Environ Microbiol 69: 3932–3937.
Nyholm SV, Mcfall-Ngai MJ . (2004). The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol 2: 632–642.
Pinel N . (2009). Physiological and genomic insights into the biology of verminephrobacter eiseniae, a bacterial symbiont of the earthworm eisenia foetida. Ph.D. thesis, University of Washington, Seattle.
Pinel N, Davidson SK, Stahl DA . (2008). Verminephrobacter eiseniae gen. nov., sp nov., a nephridial symbiont of the earthworm Eisenia foetida (Savigny). Int J Syst Evol Microbiol 58: 2147–2157.
Ruby EG . (2008). Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6: 752–762.
Schramm A, Davidson SK, Dodsworth JA, Drake HL, Stahl DA, Dubilier N . (2003). Acidovorax-like symbionts in the nephridia of earthworms. Environ Microbiol 5: 804–809.
Schweitzer B, Huber I, Amann R, Ludwig W, Simon M . (2001). Alpha- and beta-proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl Environ Microbiol 67: 632–645.
Shepherd RW, Lindow SE . (2009). Two dissimilar N-acyl-homoserine lactone acylases of pseudomonas syringae influence colony and biofilm morphology. Appl Environ Microbiol 75: 45–53.
Simon R, Oconnell M, Labes M, Puhler A . (1986). Plasmid vectors for the genetic-analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol 118: 640–659.
Stabb EV, Ruby EG . (2003). Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol 69: 820–826.
Strom MS, Lory S . (1993). Structure-function and biogenesis of the Type-Iv Pili. Annu Rev Microbiol 47: 565–596.
Strom MS, Nunn DN, Lory S . (1993). A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Nat Acad Sci USA 90: 2404–2408.
Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T . (2001). Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun 69: 7937–7940.
Wassenaar TM, Vanderzeijst BAM, Ayling R, Newell DG . (1993). Colonization of chicks by motility mutants of Campylobacter-Jejuni demonstrates the importance of flagellin-a expression. J Gen Microbiol 139: 1171–1175.
Watson AA, Alm RA, Mattick JS . (1996). Identification of a gene, pilF, required for type 4 fimbrial biogenesis and twitching motility in Pseudomonas aeruginosa. Gene 180: 49–56.
Wilson M, Lindow SE . (1993). Interactions between the biological control agent Pseudomonas fluorescens A506 and Erwinia amylovora in pear blossoms. Phytopathology 83: 117–123.
Acknowledgements
This work was supported by the National Science Foundation Grants IOB 0951119 (to SKD) and IOB 0345049 (to DAS and SKD). Funding for GD was provided by an awarded NSF Minority Postdoctoral Fellowship (DBI 0805653). RG was funded by the REU/NSF program under amendment IOB 0619945 to NSF IOB 0345049.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dulla, G., Go, R., Stahl, D. et al. Verminephrobacter eiseniae type IV pili and flagella are required to colonize earthworm nephridia. ISME J 6, 1166–1175 (2012). https://doi.org/10.1038/ismej.2011.183
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.183
Keywords
This article is cited by
-
The draft genome of a new Verminephrobacter eiseniae strain: a nephridial symbiont of earthworms
Annals of Microbiology (2020)
-
Genome analysis of Paenibacillus polymyxa A18 gives insights into the features associated with its adaptation to the termite gut environment
Scientific Reports (2019)
-
The role of microbial motility and chemotaxis in symbiosis
Nature Reviews Microbiology (2019)
-
Insights into flagellar function and mechanism from the squid–vibrio symbiosis
npj Biofilms and Microbiomes (2019)
-
Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body
The ISME Journal (2018)