Abstract
We modified the standard ribosomal RNA-targeted fluorescence in situ hybridization (FISH) protocol by removing the fixation steps to allow recovery of unmodified nucleic acids. Using this method, hybridized cells could be visualized in two bioreactor sludges and termite hindgut samples by epifluorescence microscopy. We then targeted one bacterial and one archaeal population in the sludge samples with group-specific oligonucleotide probes using in-solution fixation-free FISH and sorted hybridized populations using fluorescence-activated cell sorting (FACS). We could show that sorted populations were highly enriched for the target organisms based on 16S rRNA gene sequencing, thus confirming probe specificity using the modified FISH protocol. This approach should facilitate subsequent genomic sequencing and analysis of targeted populations as DNA is not compromised by crosslinking during fixation.
Similar content being viewed by others
Main
Fluorescence in situ hybridization (FISH) is a powerful method for visualizing microbial populations in a community setting (Amann and Fuchs, 2008) and can be used as the basis for separating targeted populations from a multi-species background through fluorescence-activated cell sorting (FACS) (Kalyuzhnaya et al., 2006; Podar et al., 2007). Cell fixation is an accepted and reportedly integral part of the FISH protocol that serves to stabilize cell integrity and facilitate probe permeability for imaging (Amann et al., 1995). For microorganisms, this is commonly achieved with paraformaldehyde that crosslinks proteins and nucleic acids (Hayat, 2000). However, for genomic or proteomic applications, unfixed biomass is desirable to minimize possible sequencing biases introduced by crosslinking and also to avoid possible cell lysis difficulties (Wallner et al., 1997). Ribosomal RNA-targeted FISH using unfixed microbial samples has been reported for Escherichia coli, Salmonella enterica and Pseudomonas putida using specialized quenched autoligation oligonucleotide probes (Silverman and Kool, 2005; Silverman et al., 2008) that are not yet commercially available. In addition, unfixed mammalian tissue has been shown to produce better imaging results than paraformaldehyde-fixed tissue using in situ hybridization histochemistry (Dagerlind et al., 1992). The aim of this study is to determine whether fixation-free FISH using standard probes works well enough on environmental samples to allow specific sorting of targeted populations for subsequent genomic applications.
We made a simple modification to the standard FISH protocol by removing the paraformaldehyde fixation and ethanol dehydration steps, and applied broad- and narrow-specificity FISH probes to two bioreactor sludges and a termite hindgut community (Table 1). The quality of the images was good enough to clearly delineate numerous distinct morphotypes regardless of probe, fluorochrome and sample combination, suggesting that the method may be applicable to a broad range of species (Figures 1c and d). Although hybridization specificity was not systematically addressed in this initial microscopy test, we noted that the group-specific probes highlighted only the expected morphotypes (Figures 1a and b) using stringency conditions optimized for standard FISH (Table 1). Paraformaldehyde and ethanol fixation also serves to preserve environmental samples, suggesting that fixation-free FISH may only be suitable for freshly collected material. However, we found that samples stored at −80 °C, even in the absence of cryoprotectants, produced satisfactory microscopy images (Figure 1d). Although these are promising preliminary results, rigorous testing of the method will be required to determine whether fixation-free FISH can broadly substitute for fixation-based FISH methods. This includes, but is not limited to, confirmation that (1) the method works with cells with low signal intensities and Gram-positive cells (and associated permeabilization protocols), (2) probes are specific by testing with perfect and single-mismatch target sites at different hybridization stringencies and (3) quantification of targeted populations is reproducible. Until these tests are carried out, we recommend using the method only for qualitative applications with independent verification of probe specificity, for example, by marker gene sequencing.
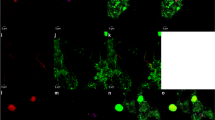
Epifluorescence microscopy images of fixation-free FISH applied to (a) sorted enhanced biological phosphorus removing (EBPR) sludge hybridized with a CAP II-specific probe highlighting the characteristic donut shape of CAP bacteria, (b) denitrifying anaerobic methane oxidizing (DAMO) sludge hybridized with archaeal-specific probes highlighting the distinctive tetrad clusters formed by ANME-2D archaea (the only archaeal members of the DAMO sludge community); the blue background is due to a concanavalin A counterstain for saccharides, (c) EBPR sludge hybridized with a broad-specificity bacterial probe highlighting several morphotypes and (d) Nasutitermes corniger hindgut community hybridized with a broad-specificity bacterial probe highlighting several morphotypes. See Table 1 for further details of the probes and hybridization conditions used.
To apply fixation-free FISH to FACS, we adopted an in-solution FISH protocol (Wallner et al., 1993; Kalyuzhnaya et al., 2006) that does not require the solid support (filter or slide) of the standard protocol (Manz et al., 1992; Glockner et al., 1996) or resuspension of cells removed from a solid support (Sekar et al., 2004). Two specific populations were targeted in the bioreactor communities, the bacterium Candidatus Accumulibacter phosphatis (CAP) type II (Flowers et al., 2009) and methanotrophic archaea closely related to ANME-2D (Mills et al., 2004) (Table 1). After in-solution fixation-free FISH, these populations were homogenized by sonication and flow sorted, using two-stage sorts, from their respective communities by gating cells based on their enhanced fluorescence and light scattering properties (Figure 2). We noted that unfixed cells homogenized more easily than fixed cells, perhaps because of the absence of crosslinking that may act on extracellular polymeric substances holding the cells together (Garcia Martin et al., 2006). After two sorts, the CAP-II population was enriched from 2.8% of the starting community to 97.6% (35-fold enrichment) and the ANME-2D population was enriched from 1.8% to 93.6% (52-fold enrichment; Figure 2). Multiple subsets of the second-sort hybridized populations (presumptively >93% pure), ranging in size from 102 to 105 cells, were sorted into different microtiter plate wells to confirm hybridization specificity (Supplementary Table S1). Sorted populations were alkaline lyzed and their genomic DNAs amplified using multiple displacement amplification (MDA; Supplementary Figure S1). Primers broadly targeting either bacteria or archaea were then used to PCR amplify the 16S rRNA genes from the whole genome-amplified subset populations. The putative CAP-II populations produced PCR amplicons only with the bacterial primers, and the putative ANME-2D populations produced PCR amplicons only with the archaeal primers (Supplementary Figure S2). The resulting amplicons were directly Sanger sequenced without cloning. We have previously found that mixed templates will produce sequence chromatograms with secondary peaks using broad-specificity primers (data not shown). Comparative analysis of the amplicon sequences confirmed hybridization specificity due to the presence of the FISH probe target sites (Table 1, Supplementary Figure S3) and the expected identity of the sorted populations (Supplementary Table S1). In the majority of cases the sequencing chromatograms showed negligible background signal, indicating that the populations were highly enriched for the target organisms by the two-stage sorts (Supplementary Figure S3). However, it is not feasible to sort more than ∼106 cells because of sorting volume and time constraints (Eek et al., 2007), which limits the total amount of recoverable DNA to <5 ng, and much less for smaller population sizes. With this caveat in mind, we anticipate that in-solution fixation-free FISH will allow enrichment of a range of targeted bacterial and archaeal populations for recovery of unmodified nucleic acids. This will facilitate downstream genomic applications and possibly applications requiring live material.
Flow cytometric analysis of pre- and post-sorts of (a) enhanced biological phosphorus removing (EBPR) sludge hybridized with AccII-444-Alexa488 and (b) denitrifying anaerobic methane oxidizing (DAMO) sludge hybridized with Arc915-fluorescein isothiocyanate (FITC) and Darch872-cyanine 3 (Cy3). The regions with a red outline indicate the gating used for the subsequent round of sorting (panel to right) and the values shown indicate the percentage of gated (fluorescent) cells relative to total cells.
Materials and methods
Samples
Fresh samples from denitrifying anaerobic methane oxidizing bioreactor communities were collected and pelleted by centrifugation and washed twice with phosphate-buffered saline before immediate processing. Samples from enhanced biological phosphorus removing sludge were collected from a sequencing batch reactor at the end of the aerobic phase. Note that biomass was collected from both reactors when normal bioreactor function had ceased to ensure that the target populations were low (<5%). Cells were pelleted by centrifugation and washed twice with phosphate-buffered saline and stored in 10% glycerol at −20 °C until further processing. Worker termites stored at −80 °C were prepared as previously described for hindgut segment P3 lumen fluid extraction (Warnecke et al., 2007). Enhanced biological phosphorus removing sludge and termite samples were thawed on ice before hybridization.
In-solution fixation-free FISH
Samples were washed twice with phosphate-buffered saline and resuspended in 100 μl of standard FISH hybridization buffer (Pernthaler et al., 2001). Oligonucleotide FISH probes used for the different samples and associated formamide stringencies are listed in Table 1. Cells were incubated at 46 °C for 2 to 3 h. After hybridization, cells were centrifuged for 3 min at 5000 × g, and washed twice with 100 μl standard wash buffer (Pernthaler et al., 2001) prewarmed to 48 °C. Cell pellets were resuspended in 100 μl prewarmed wash buffer and incubated at 48 °C for 20 min. Cells were concentrated by centrifugation, resuspended in 1 ml Tris-EDTA (pH 8) buffer or phosphate-buffered saline and homogenized by sonication at an output power of 165 W for 30 s with 5 s pulse using a Sonic Dismembrator (Fisher Scientific, Pittsburgh, PA, USA; Model 550: output power 550 W, frequency 20 kHz) or Branson Sonifier 250 (Branson Ultrasonics Corp., Danbury, CT, USA).
Fluorescence-activated cell sorting
Flow sorting of the CAP-II population from enhanced biological phosphorus removing sludge was performed using an Influx (Cytopeia Inc., Seattle, WA, USA) cell sorter in two stages to reduce final sort volume. A 488-nm laser was used as the excitation source for light scattering and fluorescence. Homogenized unfixed samples were diluted to 106–107 cells ml–1 to prevent nozzle clogging, and hybridized cells with enhanced green fluorescence were detected at 531±20 nm and sorted first at high speed (25 000 cells s–1) into 50 ml centrifuge tubes on ice. Cells from the initial sort were then resorted at lower speed (10 000 cells s–1) into PCR tubes (102–105 cells per tube). Flow sorting of ANME-2D archaea from denitrifying anaerobic methane oxidizing sludge was performed using a BD FACSAria (BD, Franklin Lakes, NJ, USA). A 488-nm laser was used as the excitation source for light scattering and fluorescence. Hybridized cells were sorted at a speed of 20 000 cells s–1 into 2 ml Eppendorf tubes (104 cells per tube) based on forward scatter and fluorescence intensity. The effectiveness of population enrichment was verified by viewing aliquots of sorted cells using epifluorescence microscopy.
Whole genome amplification and 16S rRNA-based molecular identification
Genomic DNAs were released from sorted cells using alkaline lysis and amplified using the REPLI-g Ultrafast Mini kit (for the CAP-II population) (QIAGEN Inc., Valencia, CA, USA) or the Illustra GenomiPhiV2 DNA amplification kit (for the ANME-2D population) (GE Healthcare, Piscataway, NJ, USA) in a 20 μl volume for 2 h (GenomiPhi) or 12 h (REPLI-g) at 30 °C. The reaction was terminated by heating at 65 °C for 3 min before cooling to 4 °C. 16S rRNA genes were PCR-amplified from MDA products using broad-specificity primers: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) for the bacterial CAP-II population or Arc8f (5′-TCCGGTTGATCCTGCC-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for the archaeal ANME-2D population. Bacterial amplicons were directly Sanger sequenced, without cloning, using a 27F and/or 530F (5′-GTGCCAGCAGCCGCG-3′) primer and archaeal amplicons sequenced using a 1391R (5′-GACGGGCGGTGWGTRCA-3′) primer. Amplicon sequences were compared against the publicly available databases to confirm population identity.
References
Amann R, Fuchs BM . (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Micro 6: 339.
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M . (1999). The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444.
Dagerlind A, Friberg K, Bean AJ, Hokfelt T . (1992). Sensitive mRNA detection using unfixed tissue: combined radioactive and non-radioactive in situ hybridization histochemistry. Histochemistry 98: 39–49.
Eek KM, Sessions AL, Lies DP . (2007). Carbon-isotopic analysis of microbial cells sorted by flow cytometry. Geobiology 5: 85–95.
Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD . (2009). Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘Candidatus Accumulibacter’ clades. Environ Microbiol Rep 1: 583–588.
Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC et al. (2006). Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 24: 1263–1269.
Glockner FO, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K et al. (1996). An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol 19: 403–406.
Hayat MA . (2000). Principles and Techniques of Electron Microscopy: Biological Applications. Cambridge University Press: Cambridge.
Kalyuzhnaya MG, Zabinsky R, Bowerman S, Baker DR, Lidstrom ME, Chistoserdova L . (2006). Fluorescence in situ hybridization-flow cytometry-cell sorting-based method for separation and enrichment of type I and type II methanotroph populations. Appl Environ Microbiol 72: 4293–4301.
Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH . (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria—problems and solutions. Syst Appl Microbiol 15: 593–600.
Mills HJ, Martinez RJ, Story S, Sobecky PA . (2004). Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities from Gulf of Mexico cold-seep sediments. Appl Environ Microbiol 70: 5447–5458.
Pernthaler J, Glöckner F-O, Schönhuber W, Amann R . (2001). Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. In: HP John (ed). Methods in Microbiology. Academic Press: London, pp 207–226.
Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA et al. (2007). Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol 73: 3205–3214.
Raghoebarsing AA, Pol A, Tvan de Pas-Schoonen K, Smolders AJ, Ettwig KF, Rijpstra WI et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440: 918–921.
Sekar R, Fuchs BM, Amann R, Pernthaler J . (2004). Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol 70: 6210–6219.
Silverman AP, Abe H, Kool ET . (2008). Quenched autoligation probes. Methods Mol Biol 429: 161–170.
Silverman AP, Kool ET . (2005). Quenched autoligation probes allow discrimination of live bacterial species by single nucleotide differences in rRNA. Nucleic Acids Res 33: 4978–4986.
Stahl DA, Amann R . (1991). Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons Ltd: Chichester, UK. pp 205–248.
Wallner G, Amann R, Beisker W . (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14: 136–143.
Wallner G, Fuchs B, Spring S, Beisker W, Amann R . (1997). Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol 63: 4223–4231.
Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al. (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–565.
Acknowledgements
We thank Norm Pace, Rudi Amann and three anonymous reviewers for insightful comments, and John Wilson, Virginia Nink and Geoff Osbourne of the Queensland Brain Institute Flow Cytometry facility for their assistance. We also thank Shihu Hu for bioreactor operation and Zhiguo Yuan for support of MFH. This work was conducted in part at the US Department of Energy Joint Genome Institute that is supported by the Office of Science of the US Department of Energy under contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Yilmaz, S., Haroon, M., Rabkin, B. et al. Fixation-free fluorescence in situ hybridization for targeted enrichment of microbial populations. ISME J 4, 1352–1356 (2010). https://doi.org/10.1038/ismej.2010.73
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.73
Keywords
This article is cited by
-
FISH-TAMB, a Fixation-Free mRNA Fluorescent Labeling Technique to Target Transcriptionally Active Members in Microbial Communities
Microbial Ecology (2022)
-
Coniochaeta fungus benefits from its intracellular bacteria to form biofilm and defend against other fungi
Archives of Microbiology (2021)
-
A pipeline for targeted metagenomics of environmental bacteria
Microbiome (2020)
-
Microbial single-cell omics: the crux of the matter
Applied Microbiology and Biotechnology (2020)
-
Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria
Scientific Reports (2019)