Abstract
Bacteria associated with the onset of type 1 diabetes in a rat model system were identified. In two experiments, stool samples were collected at three time points after birth from bio-breeding diabetes-prone (BB-DP) and bio-breeding diabetes-resistant (BB-DR) rats. DNA was isolated from these samples and the 16S rRNA gene was amplified using universal primer sets. In the first experiment, bands specific to BB-DP and BB-DR genotypes were identified by automated ribosomal intergenic spacer analysis at the time of diabetes onset in BB-DP. Lactobacillus and Bacteroides strains were identified in the BB-DR- and BB-DP-specific bands, respectively. Sanger sequencing showed that the BB-DP and BB-DR bacterial communities differed significantly but too few reads were available to identify significant differences at the genus or species levels. A second experiment confirmed these results using higher throughput pyrosequencing and quantitative PCR of 16S rRNA with more rats per genotype. An average of 4541 and 3381 16S rRNA bacterial reads were obtained from each of the 10 BB-DR and 10 BB-DP samples collected at time of diabetes onset. Nine genera were more abundant in BB-DP whereas another nine genera were more abundant in BB-DR. Thirteen and eleven species were more abundant in BB-DP and BB-DR, respectively. An average of 23% and 10% of all reads could be classified at the genus and species levels, respectively. Quantitative PCR verified the higher abundance of Lactobacillus and Bifidobacterium in the BB-DR samples. Whether these changes are caused by diabetes or are involved in the development of the disease is unknown.
Similar content being viewed by others
Introduction
Type 1 diabetes (T1D) results from a complex interplay between varying degrees of genetic susceptibility and environmental factors. A trio of interacting facets that create a ‘perfect storm’ for diabetes development have been proposed including (1) an aberrant intestinal microbiota, (2) a ‘leaky’ intestinal mucosal barrier and (3) altered intestinal immune responsiveness (Vaarala et al., 2008). The first of these, an aberrant intestinal microbiota, the focus of this investigation, could readily lead to mucosal barrier dysfunction and altered intestinal immune responsiveness.
Four lines of evidence support importance of microbes in the development of autoimmunity and subsequent insulin-dependence in rodent models for T1D. First, probiotic treatment of non-obese diabetic (NOD) mice prevents the onset of T1D (Calcinaro et al., 2005; Yadav et al., 2007). Similarly, a low-fat diet with Lactobacillus strains reduced diabetes in rats (Matsuzaki et al., 1997). Second, antibiotics can prevent T1D in bio-breeding diabetes-prone (BB-DP) rats (Brugman et al., 2006) and in NOD mice (Schwartz et al., 2007). Third, the incidence of diabetes in NOD mice increases in a germ-free environment (Suzuki et al., 1987; Wicker et al., 1987). And fourth, Freund's adjuvant, which contains mycobacteria, also protects NOD mice and the BB-DP rats against diabetes (McInerney et al., 1991; Sadelain et al., 1990a, 1990b).
The reduction of diabetes in an animal model fed antibiotics suggests that there are bacteria that are involved in inducing disease. The reduction of diabetes through the feeding of probiotic strains or injection with Freund's adjuvant suggests that other bacteria may be involved in counteracting the action of those bacteria that reduce disease or they may help in triggering an immune response against the harmful bacteria.
These results encouraged us to do a culture-independent analysis of the bacteria in fecal samples collected from bio-breeding diabetes-resistant (BB-DR) and BB-DP rats in two separate experiments. The techniques used included automated ribosomal intergenic spacer analysis (ARISA), DNA Sanger sequencing and high-throughput pyrosequencing and quantitative PCR (qPCR).
Materials and methods
Animals, stool sampling and DNA extraction
Two experiments were conducted with two independent sets of rats. In experiments 1 and 2, three and ten rats of each genotype were used, respectively. Stool samples were collected at 20, 30 and 70 days after birth. The genotypes were the BB-DR and BB-DP rats. In BB-DP rats, the onset of diabetes begins at 70 days. For animal housing, Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) standards were used with four male or five female rats per cage under pathogen-free conditions. BB-DR and BB-DP rats were kept in separate cages but all were in the same room at the same temperature and light. All rats received the same water and food. All animals were put into an individual cage for the stool collection for about 4–5 h to eliminate contamination from the stool of other animals. Rats were housed and samples were obtained from Biomedical Research Models Inc. (Worcester, MA, USA).
After storage at −20 °C until DNA extraction, bacterial DNA was isolated from the stool samples using the FastDNA Kit (Qbiogene Inc., Carlsbad, CA, USA). After the DNA extraction, samples were purified with the DNeasy Tissue kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions.
ARISA analysis
Bacterial community composition was assessed by ARISA, a culture-independent technique for constructing bacterial community fingerprints based on the length heterogeneity of the intergenic transcribed spacer region of bacterial rRNA operons (Fisher and Triplett, 1999; Bosshard et al., 2000). ARISA was modified by separating PCR products on a chip with an Agilent Bioanalyzer 2100 (Agilent Technology, Santa Clara, CA, USA). In this study, ARISA profiles were assumed to be indicative of bacterial community composition, and differences in ARISA profiles were assumed to reflect variation in the composition of the respective bacterial communities.
PCR was performed with the GoTaq PCR core system (Promega, Madison, WI, USA). The mixtures contained 5 μl of 10 × PCR buffer, 200 μM dNTPs, 100 μM of each primer, 2.5 U of Taq polymerase and approximately 100 ng of DNA template in a final volume of 50 μl. The primers used were S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 (Ranjard et al., 2001). Reaction mixtures were held at 94 °C for 3 min, followed by 30 cycles of amplification at 94 °C for 45 s, 55 °C for 1 min and 72 °C for 2 min and a final extension of 72 °C for 7 min.
ARISA PCR products were purified with the QIAquick PCR Purification Kit (Qiagen). The amplification products fragments were then resolved by on-chip gel electrophoresis with an Agilent 2100 Bioanalyzer and the DNA LabChip Kit 7500 (Agilent Technology). Size standards were also resolved in separate wells to estimate the size of each PCR product. The data were translated into gel-like images where peaks from the electropherograms are translated to appear as bands on a gel (Figure 1). For each ARISA data set, the size, number and area of peaks in the electropherograms were used to compare samples. Area peaks were standardized by dividing each single area peak by the total area of the peaks in the same sample.
Gel-like image generated by the bioanalyzer. The first and last columns are the reference DNA 7500 ladder. Base pair sizes are indicated adjacent to the ladder. Samples 1–3 represent the automated ribosomal intergenic spacer analysis (ARISA) profiles for the intestinal tract of bio-breeding diabetes-prone (BB-DP) rats and samples 4–6 represent bio-breeding diabetes-resistant (BB-DR) rats' stool samples at 60 days of age. The lowermost (50 bp) and the uppermost (10 380 bp) bands represent the markers used to align the ladder data with data from the sample wells. The boxes represent dominant bands unique to both groups and were extracted from the gel for further sequencing. The stool samples used in the ARISA come from experiment 1.
To assess the degree of similarity among the samples, bacterial diversity and richness were calculated based on ARISA profiles. The bacterial diversity was estimated by using Shannon–Weaver index (H=sum (Pi ln(Pi)), where Pi is the number of a given species divided by the total number of species observed. The richness was estimated by using Margalef's index (d=(S−1)/log N), where S is the total number of species and N is the total number of individuals in the sample that provides a measure of species richness that is roughly normalized for sample size.
The Bray–Curtis similarity index was calculated to assess the degree of similarity among the samples and produce a similarity matrix. The resulting matrices with pairwise similarities were used to group samples that represented similar bacterial community composition. Hierarchical clustering was calculated by using complete linkage algorithm and the results were represented by a dendrogram with the x axis representing the full set of samples and the y axis defining a similarity level at which two samples were considered to have fused. All data analyses for the ARISA bands were conducted using the software PRIMER 6 version 6.1.9 (PRIMER-E Ltd, Luton, UK).
To identify the bacteria represented within specific ARISA bands, the PCR products were resolved on a 1.2% agarose gel and the appropriate bands excised. The bands were purified using the QIAEX II Gel Extraction Kit (Qiagen). The purified fragments were cloned into a TOPO TA Cloning (Invitrogen, Carlsbad, CA, USA). Plasmids were purified with the QIAprep Spin Miniprep Kit (Qiagen) and the DNA fragments were cycle sequenced in both directions with T7 and T3 primers using DYEnamic ET Terminator Cycle Sequencing Kit (GE Healthcare), on a PTC-200 Thermocycler (Bio-Rad) and run on a 96-well MegaBACE 1000 Capillary Sequencer (GE Healthcare).
Vector sequence present in each sequence was eliminated using VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html). After elimination of potentially chimeric sequences, the nucleotide sequences were compared to all sequences in the NCBI database using MegaBLAST (Altschul et al., 1997). Bacterial taxonomic affiliations were assigned based on the closest NCBI match. The criteria to assign a sequence to its closest relative were based on the best E-value (greater than 1E−50) and on the best bit-score (greater than 200). Sequences that did not match this criterion remained unclassified.
16S rRNA gene amplification, cloning and sequencing
A fragment of 16S rRNA gene was amplified from the 60-day samples to confirm that the bacterial communities in both rat lines had differentiated by that time. To amplify the 16S rRNA gene fragments, primers 787f and 1492r were chosen (Roesch et al., 2007). The PCR conditions used were 94 °C for 2 min, 30 cycles of 94 °C, 45 s denaturation; 55 °C, 45 s annealing and 72 °C, 1 min extension, followed by 72 °C for 6 min. The PCR products were cleaned, cloned into TOPO TA Cloning, purified, sequenced and sequences trimmed as described above. The sequences were aligned using ClustalX 1.83 (Thompson et al., 1997). Both weighted and unweighted UniFrac (Lozupone et al., 2006) were used to perform a principal coordinates analysis (PCA) to find clusters of small groups of samples. The 16S rRNA gene sequences were classified using a MegaBLAST search using the RDP II database. GenBank accession numbers EU812993 to EU814325 have been assigned to the DNA sequences obtained in this work.
16S rRNA gene amplification and pyrosequencing
A fragment of 16S rRNA gene from the V9 region was amplified from the DNA extracted from the stool samples. We amplified the 16S rRNA gene fragments using the primers 787f and a modification of 1492r (Roesch et al., 2007). The primers were attached to the 454 Life Sciences primer B and A (454 Life Sciences Corp., Branford, CT, USA). The 454 run were multiplexed to obtain 16S rRNA sequences from 20 samples simultaneously. To do this, 8-base barcodes were added to the 5′-end of the reverse primers using the self-correcting barcode method of Hamady et al., (2008). For a list of primers and barcode used see Supplementary information. Six independent PCRs were performed for each sample. The PCR conditions used were 94 °C for 2 min, 25 cycles of 94 °C, 45 s denaturation; 55 °C, 45 s annealing and 72 °C, 1 min extension, followed by 72 °C for 6 min. The 6 PCR replications for each of 20 samples were combined and purified with the QIAquick PCR Purification Kit (Qiagen). The DNA concentration of the PCR product was then quantified by using on-chip gel electrophoresis with Agilent 2100 Bioanalyzer and DNA LabChip Kit 7500 (Agilent Technology). Finally, the reactions were combined in equimolar ratios to create a DNA pool that was used for pyrosequencing with primer B. The sequencing library was produced using the standard GS FLX library preparation procedure and two titration runs were performed. The average read length obtained was 215 bases, and the maximum read length was 292 bases. All reads were generated by GS FLX. All reads shorter than 100 bases were deleted from the analysis. The pyrosequences are deposited in GenBank as accession numbers FJ269364–FJ291326 and FJ291327–FJ313064 for the BB-DP sequences and FJ313065–FJ345302 for the BB-DR sequences.
Data preparation before further analysis
Initially, all pyrosequencing reads were screened for quality and length of the sequences. The ends of the sequences that presented a Phred score smaller than 20 were trimmed and those reads that are shorter than 100 bases were removed from the data set by using a program called Trim2 (Huang et al., 2003). The trimmed sequences were than screened for the 8-base barcode. A custom Perl script was written to find the barcode and generate a new file for each sample. The sequences were then relabeled accordingly to denote the original sample.
Library comparisons
For the overall comparison for significant differences among the bacterial communities evaluated, we first group the sequences from each sample into operational taxonomic units (OTUs) (cutoff value for assigning a sequence to the same group was equal to or greater than 97% similarity) using the program CD-HIT (Cluster Database at High Identity with Tolerance; Li and Godzik, 2006). Representative sequences (the longest sequence of the cluster) were chosen and merged in a single file. This file was used as input for MUSCLE (Edgar, 2004), which built a guide tree using UPGMA (unweighted pair group method with arithmetic mean) agglomerative clustering method. Quantitative and qualitative bacterial diversity measures were done using UniFrac (Lozupone et al., 2006). UniFrac analysis required a phylogenetic tree prepared using MUSCLE and the number of sequences found on each OTU in each sample. To assess the qualitative and quantitative diversity of the bacterial communities, unweighted and weighted UniFrac were used, respectively. By using UniFrac, PCA was performed to find clusters of similar groups of samples. PCA is an ordination method based on multivariate statistical analysis that maps the samples in different dimensions and reflects the similarity of the biological communities. A matrix using the UniFrac metric for each pair of environments is calculated. The distances are turned into points in space with the number of dimensions one less than the number of samples. The first three principal dimensions were used to plot a three-dimensional graph (Figure 2).
Principal coordinates analysis (PCA) depicting the qualitative (presence/absence) and quantitative (presence/absence and abundance) of the bacterial communities for the 10 stool samples each from the bio-breeding diabetes-resistant and bio-breeding diabetes-prone rats. This analysis is based on the community structures derived from Sanger sequencing of experiment 1 (A) and pyrosequencing of experiment 2 (B).
Similarity among communities based on membership and structure
To identify the organisms whose abundances differ between the BB-DP and BB-DR samples, sequence libraries were combined and each sequence was assigned to an OTU at 95% and 97% similarity by using CD-HIT (Li and Godzik, 2006). The number of sequences in each OTU found in each sample was used to construct a table with OTUs (lines) and samples (columns). This operation was performed by using a custom script written in PHP/HTML. The script uses as input the .clstr file from CD-HIT. The input data are stored in a database where these are organized in two columns. The first column represents the sample name and the second represents the OTU number. Using MySQL statements all the data contained for each OTU are collected and compared with an array that contains all of the sample names. The list grows with each new match found. After all comparisons are completed, all OTUs are phylogenetically classified.
Phylogenetic classification of 16S rRNA gene fragments
The 16S rRNA gene sequences were phylogenetically classified using BLAST searches against the RDP II database. The closest bacterial relatives were assigned according to their best matches to sequences in the database, using an E-value threshold equal to or less than 10−20 and a bit score equal to or greater than 200.
To determine whether specific bacterial genera or species differed between BB-DP and BB-DR rats, an exact χ2-test (based on 50 000 Monte Carlo iterations) was performed to get a P-value for the null hypothesis that there was no difference between the BB-DR and BB-DP rats. The exact test, which is based on permutations, is not sensitive to zero counts in the bacterial relatives. The P-values were ordered and processed to find a false discover rate cutoff of 1%.
Real-time quantification of total Bifidobacterium and Lactobacillus load
The DNA extracts were used as a template for two separated PCRs using primers first described by Delroisse et al. (2008). The primers used are F-lacto (5′-gaggcagcagtagggaatcttc-3′), R-lacto (5′-ggccagttactacctctatccttcttc-3′), F-bifido (5′-cgcgtcyggtgtgaaag-3′) and R-bifido (5′-ccccacatccagcatcca-3′). qPCRs were performed in a reaction volume of 20 μl containing 1 × FastStart SYBR Green Master Mix (Roche Diagnostics, Indianapolis, IN, USA), 200 nM each forward and reverse primers and 5 ng of DNA extracted from the stool samples. Amplification and detection of DNA were performed with the iCycler Detection System (Bio-Rad) with optical grade 96-well PCR plates and optical film. The reaction conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 62 °C for 1 min. Data analysis was conducted with the software supplied by Bio-Rad. Purified genomic DNA in the range 10 fg to 1 ng of Lactobacillus reuteri were used as the standard for determining the amount of Lactobacillus or Bifidobacterium DNA by real-time PCR. Using L. reuteri DNA as a standard for both genera is appropriate given that both have similar genome sizes. DNA concentrations were determined with the NanoDrop spectrophotometer.
Calculation of bacterial cell numbers following qPCR
The conversion of the amount of Lactobacillus and Bifidobacterium DNA into cell numbers in the stool samples was determined as described by Byun et al. (2004). In this method, an average genome size of 2.2 Mb is assumed along with similar 16S rRNA gene copy numbers (Klaenhammer et al., 2002; Makarova et al., 2006). With these parameters, each cell is assumed to contain approximately 2.4 fg of DNA (Byun et al., 2004).
Results
Direct sequence analysis of ARISA bands: experiment 1
Two dominant peaks, 600–640 bp in size, were found in all the BB-DP samples but not in the BB-DR samples (Figure 1). A dominant 370-bp peak was also found in all BB-DR samples, which was not present among the stool samples from BB-DP rats (Figure 1). All three of these bands were excised from agarose gels and sequenced. A total of 247 sequences were obtained for the BB-DP-specific 600- and 640-bp bands. The sequencing analysis showed that these bands were derived from strains of Bacteroides, Xanthomonas and Acinetobacter. The genera Bacteroides made up 44.9% of the sequences whereas Xanthomonas and Acinetobacter were found in 15.8% and 14.5% of the sequences tested, respectively. A total of 266 sequences were obtained for the 370-bp band that was BB-DR specific. Lactobacillus strains were the source of 92.8% of the sequences with the remaining sequences derived from Clostridium, Flexibacter and Porphyromonas. These results suggested that Lactobacillus may be more common in BB-DR than in BB-DP and that Bacteroides may be more common in BB-DP.
Comparison of the bacterial communities in BB-DR and BB-DP based on 16S rRNA library comparison: experiment 1
An average of 138 16S rRNA sequences were obtained from the six stool samples from experiment 1. These sequences were aligned to prepare a distance matrix by calculating pairwise UniFrac values (Lozupone et al., 2006) for each BB-DP and BB-DR sample at 60 days of age. PCA of the matrix was constructed using UniFrac. As the number of OTUs are correlated with the amount of sampling effort (Hughes et al., 2001; Roesch et al., 2007), a simple comparison of the number of OTUs between groups can lead to misinterpretations due to undersampling or due to variability between individuals rather than between groups. To avoid this problem, quantitative and qualitative bacterial diversity measures were calculated by using PCA. This approach compares the communities for significant differences using phylogenetic information and multivariate statistical techniques for finding the most important axes along which the samples vary. In this study, PCA was also used to find clusters of samples that represent similar bacterial communities (Figure 2).
PCA showed that the BB-DP communities were far more similar to each other than they were to any of the BB-DR communities (Figure 2a). The BB-DP and BB-DR communities differed at the 1% level of confidence as measured using either weighted or unweighted UniFrac. Thus, the composition of the bacterial community differed whether or not the abundance of taxa was considered.
Analysis of bacterial community composition in BB-DP and BB-DR samples: experiment 2
The results of experiment 1 encouraged a second experiment with more replicates per genotype and higher throughput 16S rRNA sequencing. The first analysis of the pyrosequencing data was to determine whether the bacterial communities found in the 20 rats at 70 days after birth differed significantly between the BB-DR and BB-DP rats. This was done by PCA as described above in experiment 1 (Figure 2b).
The weighted and unweighted UniFrac analyses showed that the bacterial community composition in the stool of BB-DP and BB-DR rats was significantly different at the 1% level of confidence. The Shannon–Weaver and richness diversity indices were calculated for each time point in experiment 2 using the ARISA profiles (Figure 3). The diversity indices did not change significantly over time in the BB-DR samples but did decline by 70 days in the BB-DP samples. There were no significant differences in diversity between the 20- and 30-day samples in BB-DP or BB-DR.
Shannon–Weaver and richness diversity indices calculated for the three time points after birth when stool was collected. Circles and squares represent the BB-DP and BB-DR samples, respectively. Open symbols represent the richness index (d) whereas closed symbols depict the Shannon–Weaver (H′) indices. Indices were calculated using automated ribosomal intergenic spacer analysis (ARISA) data from the experiment 2 samples.
Identification of the bacteria that vary between the BB-DP and BB-DR samples: experiment 2
Having found statistically significant differences between bacterial communities in BB-DP and BB-DR rats (Figures 1 and 2), the next step for us was to identify those bacterial genera and species that were responsible for the differences observed. To determine the types of intestinal bacteria associated with TD1, we assigned the 16S rRNA sequences to closest bacterial relatives according to their best matches to sequences of known organisms by using BLAST search (Altschul et al., 1997). Twenty 16S rRNA libraries were obtained by multiplex pyrosequencing and these ranged in size from 1261 to 7997 sequences (Hamady et al., 2008; Supplementary Table S1). Sequences within each library were compared and OTUs were identified using 95% or 97% similarity as criteria for assigning sequences to approximate the same genus or species, respectively. For the BB-DP samples, the number of OTUs at the 97% similarity level varied from 327 to 1210 with an average of 748. For the BB-DR samples, the number of OTUs varied from 270 to 1689 with an average of 724.
The proportion of total reads that could be assigned to known genera was 22.33% and 23.65% of the BB-DP and BB-DR reads, respectively, using the 95% similarity level to define a genus. The proportion of total reads that could be assigned to known species was 12.06% and 13.23% of the BB-DP and BB-DR reads, respectively, using the 97% similarity level to define a species.
The bacterial communities were compared at the genus and species level with 74 bacterial genera and 124 bacterial species identified as inhabitants of the rat stool tested. To test which genera or species were different between BB-DR and BB-DP rats, an exact χ2-test showed that 24 bacterial species and 18 bacterial genera differed in abundance at the 1% level of confidence between BB-DP and BB-DR samples (Tables 1 and 2). Those species and genera that did not differ are also presented (Supplementary Tables S2 and S3). The most abundant genera found in these samples were Clostridium and Bacteroides. The abundance of the Bacteroides differed significantly between the BB-DP and BB-DR samples whereas the abundance of Clostridium did not change (Supplementary Table S2). Five species of Clostridium were more abundant in BB-DP whereas one species was more abundant in BB-DR (Table 2).
On the basis of the exact χ2-test, nine genera were found to be statistically significantly higher abundant in the BB-DP samples but in statistically significantly lower abundant or absent in the BB-DR samples (Table 1). These genera were Bacteroides, Eubacterium, Halothermothrix, Ruminococcus, Anaerostipes, Mucispirillum, Butyrivibrio, Pediococcus and Sporobacter. Of these, Bacteroides, Eubacterium and Ruminococcus were the most abundant in the BB-DP samples with 6.73%, 4.00% and 2.30%, respectively, of the total number of sequences.
In BB-DR rats, nine bacterial genera were found in statistically greater numbers than in the BB-DP samples (Table 1). These genera include Bifidobacterium, Lactobacillus, Prevotella, Pseudobutyrivibrio, Spiroplasma, Proteiniphilum, Streptococcus, Turicibacter and Bryantella. Of these, the most abundant were Lactobacillus, Bryantella, Bifidobacterium and Turicibacter.
The physiology of bacterial species within a genus can vary. Thus, species differences within each genus between the two rat genotypes were considered an important component of these analyses. At the 1% level of confidence, 24 bacterial species differed in abundance between the two rat genotypes (Table 2). Among them, 11 were more abundant in BB-DR and 13 were more abundant in BB-DP. Some of these differences were within a genus that did not differ between the two rat genotypes. For example, although the number of reads of Clostridium did not differ between the two treatments, six species of Clostridium did differ. Of these, five species were more abundant in BB-DP whereas one species was more abundant in BB-DR (Table 2).
At the genus level, Bacteroides strains were more prevalent in BB-DP than in BB-DR rats. However, at the species level, strains of B. capillosus, B. vulgatus and B. splanchnicus were more common in BB-DP whereas strains of B. acidifaciens and B. massiliensis were more common in BB-DR. Four Lactobacillus species were more common in BB-DR samples but an unidentified clade of Lactobacillus was more common in BB-DP samples.
Verification of the pyrosequencing results: quantification of Lactobacillus and Bifidobacterium in the BB-DP and BB-DR samples, experiment 2
In attempt to verify the accuracy of the results obtained by using pyrosequencing, qPCR was performed to quantify the abundance of two bacterial genera (Lactobacillus and Bifidobacterium) that are more common in BB-DR than in BB-DP samples. The Ct values obtained were converted into cell number and the averaged number of bacterial cells for the BB-DR and BB-DP samples (Figure 4). Both genera were more common in BB-DR than in BB-DP samples confirming the pyrosequencing results.
Log of the number of Lactobacillus and Bifidobacterium cells per 5 ng of DNA from bio-breeding diabetes-resistant (BB-DR) and bio-breeding diabetes-prone (BB-DP) stool samples. (a) Experiment 1 (three stool samples per genotype). (b) Experiment 2 (10 stool samples per genotype). The standard error about the mean is depicted in the error bar about the data columns.
The expression of bacterial abundance in qPCR experiments as cell numbers is well documented and accepted in the literature (Byun et al., 2004; Martin et al., 2006). In addition, more, an excellent correlation has been shown between L. sakei counts estimated by real-time PCR and L. sakei counts on MRS plates (Figure 2, Table 3; Martin et al., 2006). In this report we used the same approach. Briefly, the conversion to cell numbers is based on a calibration curve in which a small fragment from the 16S rRNA gene specific to Lactobacillus or to Bifidobacterium is amplified. To this end, chromosomal DNA from either L. reuteri (wild-type isolate) or Bifidobacterium sp. (wild-type isolate) were quantified and serial dilutions were made. Different concentrations from 50 fg to 5 ng (20–2 × 106 genome equivalents) were used as a standard curve in the qPCR experiments.
A large number of sequences not classified to the genus level
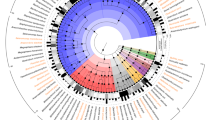
Most of the pyrosequences obtained could not be classified at the genus or species levels. Of those, 252 OTUs differed significantly between BB-DP and BB-DR. Of those, 139 were significantly more abundant in BB-DR whereas 113 were significantly higher in BB-DP. These organisms represented 24.4% and 21.4% of all reads in BB-DP and BB-DR, respectively. Of the 252 OTUs, 245 could be classified into 5 bacterial families: Clostridiaceae, Lachnospiraceae, Ruminococcaceae, Porphyromonadaceae and Prevotellaceae (Figure 5, Table 3). Of the 41 OTUs that differed between the two rat genotypes in the Clostridiaceae, 95% were more abundant in BB-DP than in BB-DR. A similar trend was found in the Ruminococcaceae, where of the 56 OTUs, 67.9% were more abundant in BB-DP than in BB-DR. Conversely, of the 119 OTUs that differed between the two rat genotypes in the Lachnospiraceae, 77% were more abundant in BB-DR than in BB-DP. The OTUs from Porphyromonadaceae and Prevotellaceae were also more likely to be in higher numbers in BB-DR than in BB-DP.
Family-level phylogenetic classification of those OTUs that could not be classified at the genus or species levels. Red branches depict 16S rRNA sequences from bio-breeding diabetes-prone (BB-DP) rats and green branches depict sequences from bio-breeding diabetes-resistant (BB-DR) rats. Branches in black depict known sequences from bacterial isolates. A list of bacterial isolates that were aligned with the sequences obtained from this study can be seen in the supplementary material. Sequences were aligned by using NAST (DeSantis et al., 2006), the aligned sequences and its respective nearest isolates were uploaded in MEGA 4 (Tamura et al., 2007) for conduction of the phylogenetic analysis. The evolutionary history was inferred using the neighbor-joining method and the evolutionary distances were computed using the maximum composite likelihood method. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). Striking taxonomic trends were observed with the Clostridiaceae and Ruminococcaceae more prevalent in BB-DP whereas the Lachnospiraceae, Porphyromonadaceae and Prevotellaceae were more common in BB-DR.
Discussion
The presence of a commensal intestinal microbiota in infancy is critical for numerous physiological processes including growth, angiogenesis, optimization of nutrition and stimulation of various arms of the innate and adaptive immune systems (Hooper and Gordon 2001; Stappenbeck et al., 2002; Hooper 2004; Mazmanian et al., 2005). However, similar studies in T1D are limited. In rodent models of T1D, the disease is likely to develop under germ-free conditions (Suzuki et al., 1987; Wicker et al., 1987; Greiner et al., 2001; Mordes et al., 2004). BB-DP rats subjected to cesarean derivation develop accelerated disease (Like et al., 1991). In terms of using such information to proactively modulate diabetes formation, the antibiotic treatments to BB-DP rats after weaning (Brugman et al., 2006) prevents diabetes, whereas in our own efforts using the NOD mouse, a decreased frequency of T1D was observed with the administration of doxycycline (Schwartz et al., 2007). The specific mechanisms of how such therapies modulate disease are unclear, but it is clear that changes in the microbiota affect the development of autoimmune diabetes in both animal models. Supporting this view, oral probiotic administration to NOD mice was noted to induce IL-10 (that is, an anti-inflammatory cytokine) production and to prevent the development of T1D (Calcinaro et al., 2005). Interest exists on intestinal microbiota composition, a metric that has previously relied almost exclusively on the quantitative cultures from feces.
In this work, a thorough, culture-independent examination of the diversity of bacteria in the stool of BB-DP and BB-DR rats was performed just before the onset of diabetes by a variety of culture-independent approaches. The results of all approaches were in agreement that certain bacterial species known for their probiotic activities are more common in BB-DR than in BB-DP rats. The results were verified with two genera using qPCR.
In these 16S rRNA libraries, close relatives of 74 genera were identified. Of those, 18 genera showed higher abundance in one rat genotype versus the other. Of the nine genera with higher abundance in BB-DR, three genera, Bifidobacterium, Lactobacillus and Pseudobutyrivibrio, have representatives with known probiotic activity (Silva et al., 1987; Vandenbergh 1993; Jack et al., 1995; Bernet-Camard et al., 1997; Lorca et al., 2001; Reid et al., 2003; Leahy et al., 2005; Ohkawara et al., 2005; Fukuda et al., 2006; Ohkawara et al., 2006; Salminen et al., 2006; Čepeljnik et al., 2003, 2007). These observations from pyrosequencing were verified by qPCR of Bifidobacterium and Lactobacillus. These results also confirmed the BB-DR-specific ARISA band identified in experiment 1 as Lactobacillus. These probiotic bacteria may prevent the growth of probiotic-sensitive strains that either cause a leaky gut epithelium or cause an altered immune response against gut microbiota.
Of the nine genera in higher abundance in BB-DP, none is known to have probiotic activity. As expected in stool samples, there are many genera that are strict anaerobes and these genera are found in BB-DR and BB-DP samples. A halophilic genus, Pontibacillus, is found in much higher numbers in BB-DP samples. These observations are consistent with previous work where feeding probiotics or antibiotics to either NOD mice or BB-DP rats prevented diabetes (Matsuzaki et al., 1997; Calcinaro et al., 2005; Brugman et al., 2006; Yadav et al., 2007).
However, there is value in looking beyond the genus level. Species-level differences reveal changes not seen at the genus level. Perhaps the most dramatic example of this is Clostridium. At the genus level, Clostridium abundance does not differ between BB-DR and BB-DP. However, five species of Clostridium are higher in BB-DP than in BB-DR. Only one Clostridium species, C. hylemonae, is higher in BB-DR than in BB-DP. However, of the six species, C. hylemonae appears to be by far the least abundant of these six species. Twenty-one other species of Clostridium were identified in these samples but they did not differ between the two genotypes (Supplementary Table S3).
These data represent a first step to understanding the specific changes in gut microflora with the onset of diabetes. The next important challenge is to discover the metabolic differences that are affected by the changes in taxa. This can be assessed with a metagenomic approach. The larger challenge for the future is whether similar differences can be observed in human T1D.
Several exogenous as well as endogenous factors could affect the intestinal microbiota in these rats. The environment of these animals including food intake was the same in both BB-DP and BB-DR rats, thus minimizing its contributions to the differences in intestinal microbiota observed between the two strains. Factors other than the environment and genetic background have been shown to contribute to gut microbiota composition. For example, Mai et al. (2007) reported associations between gut microbiota and colorectal carcinogenesis and showed that individual C57BL/6J-ApcMin/+ mice that were single caged and consumed the same amounts of the same defined diet developed differences in gut microbiota that appeared associated with colorectal carcinogenesis. One endogenous factor that could be important in this work is the difference in immune status between the two groups (Elder and Maclaren 1983; Iwakoshi et al., 1999). Severe T lymphopenia is observed in BB-DP rats, irrespective of sex or the presence of diabetes, whereas numbers of B cells and serum immunoglobulin levels are normal. Another factor is that the mucosal surface of the BB-DP rat is disrupted before the development of hyperglycemia and immune-β-cell destruction (Neu et al., 2005). Which of these factors plays a primary role by affecting the others is not currently known. Our finding that the microbial communities differ between BB-DP and BB-DR rats before the time of autoimmune pancreatic destruction suggests a complex interaction between the intestinal microbiota, the intestinal surface properties and the immune system that will require additional investigation.
However, in this work, these external factors were minimized. All rats were provided the same lighting, temperature, diet, water and cage conditions. BB-DR and BB-DP rats were in separate cages with two or three rats per cage after weaning from the mother.
A striking feature of this work is the large number OTUs that differed between BB-DP and BB-DR but could not be classified to a known genus. At the family level, there were striking taxonomic trends with the Clostridiaceae and Ruminococcaceae more prevalent in BB-DP whereas the Lachnospiraceae, Porphyromonadaceae and Prevotellaceae were more common in BB-DR. This suggests a selectivity in the changes that occur in the BB-DP gut over time. Certain taxa appear to be targeted for loss over time in BB-DP. These may be under attack from the immune system or the conditions in the BB-DP gut may be less conducive to their growth. A metagenomic functional analysis of the bacteria in BB-DP and BB-DR samples would provide insight into the bacterial function suited to the altered gut environment of a diabetic animal.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Altschul S, Madden T, Schaeffer A, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Bernet-Camard MF, Lie'vin V, Brassart D, Neeser JR, Servin AL, Hudault S . (1997). The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl Environ Microbiol 63: 2747–2753.
Bosshard PP, Stettler R, Bachofen R . (2000). Seasonal and spatial community dynamics in the meromictic Lake Cadagno. Arch Microbiol 174: 168–174.
Brugman S, Klatter FA, Visser JTJ, Wildeboer-Veloo ACM, Harmsen HJM, Rozing J et al. (2006). Antibiotic treatment partially protects against type 1 diabetes in the bio-breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49: 2105–2108.
Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N . (2004). Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol 42: 3128–3136.
Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S et al. (2005). Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48: 1565–1575.
Čepeljnik T, Lah B, Narat M, Marinšek-Logar R . (2007). Adaptation of adhesion test using caco-2 cells for anaerobic bacterium Pseudobutyrivibrio xylanivorans, a probiotic candidate. Folia Microbiol (Praha) 52: 367–373.
Čepeljnik T, Zorec M, Kostanjsek R, Nekrep FV, Marinsek-Logar R . (2003). Is Pseudobutyrivibrio xylanivorans strain Mz5T suitable as a probiotic? An in vitro study. Folia Microbiol (Praha) 48: 339–345.
Delroisse JM, Boulvin AL, Parmentier I, Dauphin RD, Vandenbol M, Portetelle D . (2008). Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol Res 163: 663–670.
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al. (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Elder ME, Maclaren N . (1983). Identification of profound peripheral T lymphocyte immunodeficiencies in the spontaneously diabetic BB rat. J Immunol 130: 1723–1731.
Fisher MM, Triplett EW . (1999). Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol 65: 4630–4636.
Fukuda S, Suzuki Y, Murai M, Asanuma N, Hino T . (2006). Isolation of a novel strain of Butyrivibrio fibrisolvens that isomerizes linoleic acid to conjugated linoleic acid without hydrogenation, and its utilization as a probiotic for animals. J Appl Microbiol 100: 787–794.
Greiner DL, Rossin AA, Mordes JP . (2001). Translating data from animal models into methods for preventing human autoimmune diabetes mellitus: caveat emptor and primum non nocere. Clin Immunol 100: 134–143.
Hamady M, Walker J, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
Hooper LV . (2004). Bacterial contributions to mammalian gut development. Trends Microbiol 12: 129–134.
Hooper LV, Gordon JI . (2001). Commensal host-bacterial relationships in the gut. Science 292: 1115–1118.
Huang X, Wang J, Aluru S, Yang SP, Hillier L . (2003). PCAP: a whole-genome assembly program. Genome Res 13: 2164–2170.
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM . (2001). Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67: 4399–4406.
Iwakoshi NN, Greiner DL, Rossini AA, Mordes JP . (1999). Diabetes prone BB rats are severely deficient in natural killer T cells. Autoimmunity 31: 1–14.
Jack RW, Tagg JR, Ray B . (1995). Bacteriocins from Gram-positive bacteria. Microbiol Rev 59: 171–200.
Klaenhammer T, Altermann E, Arigoni F, Bolotin A, Breidt F, Broadbent J et al. (2002). Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82: 29–58.
Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D . (2005). Getting better with bifidobacteria. J Appl Microbiol 98: 1303–1315.
Li W, Godzik A . (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659.
Like AA, Guberski DL, Butler L . (1991). Influence of environmental viral agents on frequency and tempo of diabetes-mellitus in BB/WOR rats. Diabetes 40: 259–262.
Lorca GL, Wadstrom T, de Valdez GF, Ljungh A . (2001). Lactobacillus acidophilus autolysins inhibit Helicobacter pylori in vitro. Curr Microbiol 42: 39–44.
Lozupone C, Hamady M, Knight R . (2006). UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371.
Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E et al. (2006). Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA 103: 15611–15616.
Mai V, Colbert LH, Perkins SN, Schatzkin A, Hursting SD . (2007). Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol Carcinog 46: 42–48.
Martin B, Jofré A, Garriga M, Pla M, Aymerich T . (2006). Rapid quantitative detection of Lactobacillus sakei in meat and fermented sausages by real-time PCR. Appl Environ Microbiol 72: 6040–6048.
Matsuzaki T, Nagata Y, Kado S, Uchida K, Kato I, Hashimoto S et al. (1997). Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS 105: 643–649.
Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL . (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118.
McInerney MF, Pek SB, Thomas DW . (1991). Prevention of insulitis and diabetes onset by treatment with complete Freund's adjuvant in NOD mice. Diabetes 40: 715–725.
Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL . (2004). Rat models of type 1 diabetes: genetics, environment, and autoimmunity. ILAR J 45: 278–291.
Neu J, Reverte CM, Mackey AD, Liboni K, Tuhacek-Tenace LM, Hatch M et al. (2005). Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr 40: 589–595.
Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T . (2005). Oral administration of Butyrivibrio fibrisolvens, a butyrate-producing bacterium, decreases the formation of aberrant crypt foci in the colon and rectum of mice. J Nutr 135: 2878–2883.
Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T . (2006). Effect of oral administration of Butyrivibrio fibrisolvens MDT-1 on experimental enterocolitis in mice. Clin Vaccine Immunol 13: 1231–1236.
Ranjard L, Poly LF, Lata JC, Mougel C, Thioulouse J, Nazaret S . (2001). Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl Env Microbiol 67: 4479–4487.
Reid G, Jass J, Sebulsky MT, McCormick JK . (2003). Potential uses of probiotics in clinical practice. Clin Microbiol Rev 4: 658–672.
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Sadelain MWJ, Qin HY, Lauzon J, Singh B . (1990a). Prevention of type-1 diabetes in NOD mice by adjuvant immunotherapy. Diabetes 39: 583–589.
Sadelain MWJ, Qin HY, Sumoski W, Parfeny N, Singh B, Rabinovitch A . (1990b). Prevention of diabetes in the BB rat by early immunotherapy using Freund adjuvant. J Autoimmunity 3: 671–680.
Salminen S, Benno Y, de Vos W . (2006). Intestinal colonisation, microbiota and future probiotics? Asia Pac J Clin Nutr 15: 558–562.
Schwartz RF, Neu J, Schatz D, Atkinson MA, Wasserfall C . (2007). Comment on: Brugman S (2006). Antibiotic treatment partially protects against type 1 diabetes in the bio-breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49: 2105–2108. Diabetologia 50: 220–221.
Silva M, Jacobus NV, Deneke C, Gorbach SL . (1987). Antimicrobial substance from a human Lactobacillus strain. Antimicrob Agents Chemother 31: 1231–1213.
Stappenbeck TS, Hooper LV, Gordon JI . (2002). Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA 99: 15451–15455.
Suzuki T, Yamada T, Fujimura T, Kawamura E, Shimizu M Yamashita R . (1987). Diabetogenic effects of lymphocyte transfusion on the NOD or NOD nude mouse. In: Rygaard J, Brunner N, Groem N, Spang-Thomsen M (eds). Immune-Deficient Animals in Biomedical Research, Copenhagen 1985. Karger: Basel, pp 112–116.
Tamura K, Dudley J, Nei M, Kumar S . (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higging DG . (1997). The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality tools. Nucleic Acids Res 25: 4876–4882.
Vaarala O, Atkinson MA, Neu J . (2008). The ‘perfect storm’ for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57: 2555–2562.
Vandenbergh PA . (1993). Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol Rev 12: 221–238.
Wicker LS, Miller BJ, Coker LZ, McNally SE, Scott S, Mullen Y et al. (1987). Genetic control of diabetes and insulitis in the nonobese diabetic (NOD) mouse. J Exp Med 165: 1639–1654.
Yadav H, Shalini J, Sinha PR . (2007). Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition 23: 62–68.
Acknowledgements
This work was funded by the National Institutes of Health (RO3 A142288), the Brehm Coalition and the Florida Agricultural Experiment Station. We thank Biomedical Research Models, Inc. (Worcester, MA, USA) for providing all of the fecal samples used in this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary information
Rights and permissions
About this article
Cite this article
Roesch, L., Lorca, G., Casella, G. et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3, 536–548 (2009). https://doi.org/10.1038/ismej.2009.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2009.5
Keywords
This article is cited by
-
Microbiota e patologie endocrino-metaboliche
L'Endocrinologo (2023)
-
Gut microbiota in pancreatic diseases: possible new therapeutic strategies
Acta Pharmacologica Sinica (2021)
-
Oral therapy with colonization factor antigen I prevents development of type 1 diabetes in Non-obese Diabetic mice
Scientific Reports (2020)
-
Association of prevalent vaginal microbiome of mother with occurrence of type I diabetes in child
Scientific Reports (2019)
-
Probiotic properties of Lactobacillus strains with high cinnamoyl esterase activity isolated from jeot-gal, a high-salt fermented seafood
Annals of Microbiology (2019)