Abstract
Fully understanding the biology of acid mine drainage (AMD) is central to our ability to control and manipulate its environmental impact. Although genomics and biogeochemical methods are relatively well established in the field, their combination with high-resolution imaging of intact members of microbial biofilm communities has not yet reached its full potential. Here, we used three-dimensional (3D) cryogenic electron tomography to determine the size and ultrastructure of intact ARMAN cells, a novel ultra-small archaeon, and sought evidence for their interactions with other members of its community. Within acid mine drainage biofilms, apparently free-living ARMAN cells from a deeply branched archaeal lineage have volumes of 0.009–0.04 μm3 (mean ∼0.03±0.01 μm3), only ∼92 ribosomes, yet are frequent hosts for replicating viruses. Organization within the periplasm and partitioning of ribosomes to the inner surface of the cytoplasmic membrane may be factors in size minimization. Most cells contain enigmatic tubular structures of unknown function. The low ribosome copy number per unit volume, indicative of slow growth rates and targeting of cells by diverse viruses may account for the low abundance of ARMAN cells compared with other biofilm community members. Our results provide the first 3D analysis of structural features of these novel and enigmatic cells and their interactions with at least two types of viruses. Our findings also emphasize that new biological phenomena remain to be discovered among lower abundance organisms from novel uncultivated lineages.
Similar content being viewed by others
Introduction
The minimum size for cells, and the ecological significance of ultra-small organisms, remains a controversial subject. To evaluate the question of how small viable cells can be researchers have used genome reduction and theoretical calculations of the minimum volume needed to contain essential cellular machinery (Hutchison et al., 1999; Steering Group NRC, 1999; Kolisnychenko et al., 2002; Glass et al., 2006). At least 0.008 μm3 may be required to accommodate necessary components and biosynthesize 250 proteins. This is smaller than sizes conclusively demonstrated for the archaeal parasite Nanoarchaea equitans (Huber et al., 2002) and ultra-small bacteria (Rappé et al., 2002; Sheridan et al., 2003; Malmstrom et al., 2004; Giovannoni et al., 2005; Miteva and Brenchley, 2005) but larger than cell-like objects of uncertain nature (Folk and Taylor, 2002) suggested to be important in a variety of environmental and medical phenomena (Kajander and Çiftçioglu, 1998; Goldfarb, 2004; Martel and Young, 2008).
Determination of the roles of uncultivated organisms within microbial communities is a challenging problem. Several deeply branching ARMAN (Archaeal Richmond Mine Acidophilic Nanoorganism) lineages initially defied detection in acid mine drainage (AMD) biofilms because of mismatches between their ribosomal gene sequences and those of almost all earlier described archaea. ARMAN sequences were identified in metagenomic datasets and the cells were shown to occur as relatively low abundance members of most AMD communities (Baker et al., 2006). Conventional transmission electron microscopy characterization of cells enriched by filtration suggested a cell volume of 0.006 μm3, but findings were tentative because the 3D structure was not determined. In this study, we used cryogenic electron microscopy (cryo-EM) imaging and electron tomography to accurately describe the 3D architecture of intact ARMAN cells. Microscopy results highlight features of cell organization that may be associated with small cell volumes and reveal that virus–host associations have evolved among these community members. Our results also provide the first high-resolution analysis of structural features of these novel and enigmatic cells. All specimens are directly obtained from the environment, and samples are prepared with minimum perturbation: aliquots were directly cryo-plunged and stored in liquid nitrogen until data acquisition. Although no artifacts have ever been reported as a consequence of instant freezing in the field of cryo-EM, abundant artifacts are shown to be caused by the related techniques of freeze-substitution and plastic embedding. ARMAN is no exception and our current, exhaustive work, illuminates a few typical problems in the interpretation of the first ARMAN imaging data (Baker et al., 2006).
Materials and methods
AMD samples
All samples were collected on August 22, 2007 from the Richmond Mine at Iron Mountain, in northern California. We prepared and examined biofilm samples from several sites throughout the mine. The sample used for all the 3D image data acquisition was ultraback A drift, ‘UBA’, which was found by fluorescent in situ hybridization (FISH) to have the highest abundance of ARMAN cells of all those examined. The temperature of the solution from which the sample was collected was 36 °C and the pH was 1.9.
Fluorescence microscopy
For FISH, samples were fixed in 4% paraformaldehyde for ∼2 h, washed with phosphate-buffered saline, pH 1.2 and stored in 1:1 ethanol:PBS at −20 °C within 12 h of collection. Hybridizations were performed on fixed samples as reported earlier (Ram et al., 2005), with incubation at 46 °C and washing at 48 °C for 15 min. Hybridizations were counterstained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) DNA stain. Common AMD probes were used to identify other members present in the biofilm samples including; TH1187 (all Thermoplasmatales), LF655 (all Leptospirillum spp.) and LF1252 (Leptospirillum group III, Bond et al., 2000) and ARM980 and ARM1357 (ARMAN groups, Baker et al., 2006). Examples of our FISH images and of our cryo-EM controls are provided in the online Supplementary information section.
Molecular analyses
DNA was extracted by a modified phenol–chloroform extraction method, as described by Bond et al. (2000). PCR amplification of 16S rRNA genes was done as described earlier (Baker et al., 2006). The PCR products were cloned into pCR-4 TOPO and sequences of 10 individual clones determined.
Cryo-EM specimen preparation
For cryo electron microscopy, aliquots of 5 μl were taken directly from the fresh biofilm and placed onto lacey carbon grids (Ted Pella 01881) that were pre-treated by glow-discharge. The Formvar support was not removed from the lacey carbon. The grids were manually blotted and plunged into liquid ethane by a compressed air piston, then stored in liquid nitrogen. For electron microscopy at room temperature, 5 μl aliquots were placed on glow-discharged Formvar carbon coated grids (Ted Pella 01811), then the grids were blotted and air dried.
ARMAN cell identification controls
To confirm that the cells that we examined in this study were ARMAN we homogenized and filtered the UBA sample as described earlier (Baker et al., 2006). FISH analysis of this small-cell fraction revealed that the majority of the cells were ARMAN (See Supplementary information for more details). We then imaged this sample with cryo-EM. This revealed that the ARMAN cells after this filtration procedure had a similar appearance to those previously seen in filtrates (Baker et al., 2006). Therefore, the more irregular morphology as presented by Baker et al. (2006) is an artifact of the homogenization/filtration procedure (Supplementary Figures 7–9).
General imaging
Images were acquired on a JEOL–3100 electron microscope equipped an FEG electron source operating at 300 kV, an Omega energy filter, a Gatan 795 2K × 2K CCD camera and cryo-transfer stage. The stage was cooled with liquid nitrogen to 80 K for all cryogenic data sets.
To have a statistically relevant survey of cell sizes and morphologies, over 200 images were recorded using magnifications of × 36K, × 30K and × 25K at the CCD giving a pixel size of 0.83, 1.0 or 1.2 nm at the specimen, respectively. Underfocus values ranged between 8±0.5 μm and 14±0.5 μm, and energy filter widths were typically around 22±2 eV. Because our samples come directly from the environment, with no purification or cleaning steps, the thickness and transparency of the cryo-grids is highly variable. In many cases mineral aggregates, dead cells and other large objects obstructed the view of interesting cells. Slightly more than 200 images were selected for further analysis according to quality, purity and transparency of the sample. Here, 187 completely unobstructed images were used for the statistical analysis.
Electron tomography
A total of 20 tomographic data sets of ARMAN cells were acquired. These data sets included 30 ARMAN cells, 25 of which were judged of high quality. Tomographic tilt series were acquired under low-dose conditions, typically over an angular range between +65 and −65, ±5 ° with increments of 1 or 2 °. Between 70 and 124 images were recorded for each series. Eleven tilt series were acquired manually with the program Digital Micrograph (Gatan Inc.), and nine tilt series were acquired semi-automatically with the program Serial-EM (http://bio3d.colorado.edu/) adapted to JEOL microscopes.
For these tilt series data sets, all images were recorded using a magnification of × 36K, × 30K, or × 25K at the CCD giving a pixel size of 0.83, 1.0 or 1.2 nm at the specimen respectively. Underfocus values ranged between 9±0.5 and 16±0.5 μm, depending on the goal of the data set, and energy filter widths ranged between 22 to 28 eV, also depending on the data set. For all data sets the maximum dose used per complete tilt series was approximately 150 e− Å−2, with typical values of approximately 100 e− Å−2.
Software
All tomographic reconstructions were obtained with the program Imod (http://bio3d.colorado.edu/). The program ImageJ (NIH, http://rsb.info.nih.gov/ij/) was used for analysis of the two-dimensional image projections. Volume rendering and image analysis of tomographic reconstructions was done using the open source program VisIt (http://www.llnl.gov/visit) and the open source program ParaView (http://www.paraview.org/). All movies were created with the open source package ffmpeg. The ribosomes of 22 cells, the inner and outer membranes of the cell wall of four cells, and the tubular cytoplasmic organelle of two cells were segmented by hand using the program Imod. Ribosomes were also segmented using a wavelets filter built using VTK (www.kitware.com).
Ribosome processing
A total of 150 3D boxes centered on the hand-picked ribosomes were cropped from the full tomographic reconstructions of three ARMAN cells (from data sets with identical missing wedges) using the program Bsoft (http://lsbr.niams.nih.gov/bsoft/). We used a box of 48 pixels by side and a spherical mask with a diameter of 32 voxels. Individual sub-volumes were visualized and compared with the cryo-EM structure of translating E. coli ribosomes (Gilbert et al., 2004, EM DATA BANK (EMDB)/1070) with the programs Imod and ParaView. This set of ribosomes was also pre-processed with the package EMAN (http://blake.bcm.tmc.edu/emanwiki/EMAN) for centering of individual ribosomes. Alignments were computed with the program Bsoft, restricting the search in reciprocal space to account for the missing wedge. Several averages were computed for the whole set and for subsets of particles using the program Bsoft. The presence of significant heterogeneity in ARMAN cell growth stage (they are not exponentially growing in rich media, as is the case in most prior studies), and the sampling with 1.2 nm pixel size prevented a meaningful average through the entire data set. Meaningful averages of subsets of particles with similar 3D conformations were compared with the translating E. coli ribosomes (Gilbert et al., 2004, EMDB-1070) to confirm our assignment.
Results and discussion
Biofilms were collected from very acidic (pH<1) solutions containing near molar concentrations of dissolved iron and mM concentrations of Zn, Cu and As from environments within the Richmond Mine, Iron Mountain, CA, USA (for details, see Materials and methods). Samples were fixed, labeled with oligonucleotide probes that target the ribosomal RNA genes, and imaged by fluorescence microscopy. ARMAN cells were most abundant in the UBA location (pH 1.9); therefore this sample was used for ultrastructural characterization. About 30%–40% of the Archaea in the UBA sample belong to the ARMAN groups (Supplementary information); a 16S rRNA gene survey showed dominance by the ARMAN Group II lineage. Hybridization of oligonucleotides probes revealed ARMAN coexist with members of the archaeal order Themoplasmatales (Baker and Banfield, 2003) and Leptospirillum groups II and III bacteria (Materials and methods).
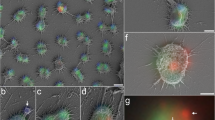
Cryo-EM observations were exclusively based on instantly frozen aliquots of fresh biofilm (Materials and methods). Within samples shown by FISH to contain an abundance of ARMAN cells (Supplementary information), ARMAN cells were distinguished from bacteria by their size and cell shape and from other archaea by the presence of a cell wall (Baker et al., 2006) (Materials and methods). Over 200 cryogenic electron microscopy (cryo-EM) images and 30 cryogenic electron tomography 3D ARMAN data sets were acquired. Nearly 200 high quality 2D projections, as described in Materials and methods, were used for measurement and statistical analyses and 25 3D reconstructions were used for analysis of the internal structure. Based on 2D images (Figure 1) and 3D information, ARMAN cell shape is generally a 3D ellipsoid (Figure 2a and Supplementary Figures 1 and 2). The three axes of an ellipsoid fitted to the inner membrane have mean values ‘a’=402 nm, ‘b’=442 nm and ‘c’=312 nm (see Supplementary Tables I and II, and Supplementary Figure 3). The volumes computed from the equatorial diameters (Supplementary Figure 3f) ranged from ∼0.009 to ∼0.062 μm3, with the mean value ∼0.03 μm3 and a s.d. of 0.01 μm3. Although the range in cell size is considerable (over two orders of magnitude in volume), it is clear that ARMAN cells can have extremely small sizes that are at or around a suggested theoretical limit for life (Steering Group NRC, 1999).
2D cryo-electron microscope images of typical ARMAN cells. (a) An individual ARMAN cell in the biofilm showing rounded shape and inner and outer membranes of the cell wall. (b) ARMAN cell that appears to be undergoing cell division. (c) Inset (magnified) shows the presence of closely spaced, high contrast features within the periplasm of another ARMAN cell (Supplementary Figure 2a). The height of the inset measures 86 nm.
Organization of ribosomes within ARMAN cells. (a) The 3D surface of a reconstructed ARMAN cell with IM and OM segmented in transparent yellow and green respectively. This is the typical oblate 3D shape of all reconstructed ARMAN cells. The ‘missing wedge effect’ was completed assuming an average curvature and the absence of singularities. A central cross section through the reconstruction is shown in gray. One voxel-thick slices of this cell are shown in Supplementary Figure 2. (b) A one voxel-thick central section of an ARMAN cell (c) 50 voxel-thick slice of the cell in (b) with the segmented IM and OM in orange and yellow respectively. Ribosomes are represented with light blue spheres drawn to scale. Low mass densities in the volume-rendered cytoplasm are in green. Tubular features, such as shown in (c), are described in Figure 3. (d) Plot of the number of ribosomes found in each 3D reconstruction as a function of the calculated volume of the cell.
Three-dimensional reconstructions of ARMAN cells in close proximity to other, considerably larger archaeal and bacterial cells and in thicker areas of the support film (where the thickness of the frozen liquid film is considerably larger than the dimensions of the ARMAN cell) confirm that the shape is an intrinsic property of the cells. Good examples are shown in Supplementary Figures 2 and 4 respectively, and in Supplementary movies 1 and 2. The oblate morphology implies cell shape determinants, probably related to the cell wall. The cell walls are smooth and uniform in thickness (∼30±2 nm wide), with conspicuous inner and outer membranes (IM, OM) and no S-layer within the dynamic range of our data (Figures 1 and 3). The periplasmic space is studded with ∼5–8 nm diameter high contrast features in close proximity to the IM (Figures 1c and 3a) and with physical connections through the IM into the cytoplasm (Figure 3a). Given that iron and sulfur oxidation are the predominant metabolisms supporting many chemoautrophs in this subsurface microbial ecosystem, it is plausible that these periplasmic elements could be cytochrome complexes, which have an estimated size of ∼40–70 kDa (Ram et al., 2005). However, this hypothesis will require future investigations with specialized techniques. Filamentous attachments associated with the cell walls as shown in Figure 3c are often observed. Their diameters, 9–10 nm, and globular structures at the IM attachment sites make them unlikely flagella candidates. We thus think they are most likely pili with ATPases around the bases.
Organization and structural features of ARMAN cells. (a) A one voxel-thick central slice through a 3D tomographic reconstruction of an ARMAN cell. Blue box: two pairs of parallel lines, clearly visible in the cytoplasm, that correspond with the cross section of a tubular structure (wall width ∼6–8 nm) also shown in 3D in Figures 2c and 3b. Ribosomes are distributed in the periphery, just inside the IM. The periplasmic space is filled with a relatively regular distribution of mass densities 5–8 nm in diameter (yellow boxes I and II). Box I shows the connections between these mass densities and the IM, whereas those in Box II lie in a different plane. (b) Two orthogonal one voxel-thick slices through a tomographic reconstruction of another ARMAN cell with the tubular structure segmented in yellow. The insets show cross sections of the tubular organelle. The diameter of the tube ranges from 58 to 62 nm and the length is ∼200 nm. (c) Average of a three voxel-thick slice through a 3D reconstruction of an ARMAN cell with a pili, diameter ∼9–10 nm.
Clearly visible within the cytoplasmic space are objects that have high contrast relative to the background. Their diameters between ∼16 and 25 nm (depending on the orientation of the slicing plane), and their shapes are consistent with their identification as ribosomes (Figures 2b and 3a, Supplementary Figures 2c and d). Careful, direct visual comparison of cross sections of these objects with the previously solved cryo-EM structure of translating E. coli ribosomes (Gilbert et al., 2004) confirmed this assignment. Additional image analysis and averaging of tomographic sub-volumes further confirmed the identification (details in Supplementary Figure 5 and Materials and methods).
In all ARMAN 3D reconstructions, ribosomes are consistently partitioned to the periphery of the cytoplasmic space, arranged in an irregular spherical pattern just inside the IM (Figure 2, Supplementary Figure 2, and Supplementary movie 1). The distribution of ribosomes has previously been investigated for Spiroplasma melliferum (Ortiz et al., 2006) by high-resolution cryogenic electron tomography In this case they were dispersed throughout most of the bacterial cytoplasmic space. Subcellular compartmentalization of ribosomes has also been proposed for Bacillus subtilis based on fluorescence microscopy (Lewis et al., 2000, and Mascarenhas et al., 2001), where it may be correlated with life cycle stages.
Given that ARMAN ribosomes are approximately the same size as bacterial ribosomes (for example, in E. Coli, Gilbert et al., 2004, and Supplementary Figure 5), it is likely that the number of ribosomal proteins/ribosome is approximately the same. However, the mean number of ribosomes per ARMAN cell (mean volume 0.03 μm3) is only 92. The number of ribosomes counted in each reconstructed ARMAN cell was plotted as a function of the cell volume (Figure 2d). The slope is very small (∼676 ribosomes per μm3) and the variance very large (∼552 ribosomes per μm3) respectively, indicating that the mean number of ribosomes per cell is an intrinsic characteristic. This linear regression predicts 745 ribosomes for a volume of 1 μm3, more than one order of magnitude less, per μm3, than in (Lewin, 1997). The much lower number of ribosomes per DNA unit length in ARMAN cells than in well-studied bacteria (∼9 ribosomes per 100 kB DNA in ARMAN vs ∼200–400 ribosomes per 100 kB DNA in E. coli, Lewin, 1997), including ultra-small bacteria (Cox, 2004; Giovannoni et al., 2005; Nicastro et al., 2006) and their peripheral distribution, suggest that protein synthesis is queued.
The relationship between the growth rate, μ, and the average number of ribosomes per cell, nR(av), is:

where βR is the fraction of ribosomes actively synthesizing protein, naa(av) is the average number of amino acids per cell (protein content), and ɛaa is the polypeptide chain elongation rate (Bremer and Dennis, 1996). Given factors such as ribosome number and genome size (which may constrain naa(av)), the growth rate of ARMAN cells is likely to be very slow (Table 1).
Tubular or cylindrical structures were identified in at least 80% of 3D reconstructions (Figures 3a and b). Tube diameters ranged from 40 to 62 nm, lengths from 20–30 to 200 nm and the wall thickness is ∼6–8 nm, comparable in diameter to a lipid bilayer. These remain an unexplained feature of ARMAN cells.
FISH and low magnification transmission electron microscopy images show ARMAN cells in the vicinity of other bacteria and archaea (Supplementary Figures 4, 7, and Movie 2) but reveal no close association or contact between them. Furthermore, we detected no anatomical feature connecting ARMAN to other cells in 3D reconstructions. Thus, we infer that ARMAN cells are free-living, to the limits of our data. Only the combination of future genomics, metagenomics and proteomics data, in addition to serially cryo-sectioned intact biofilms will further illuminate these intriguing aspects of ARMAN.
ARMAN cells have not been cultivated so it is impossible to follow their cell division processes in a synchronized, step-wise manner. Consequently, we acquired large enough data sets that all cell cycle stages normally present at a statistically significant level should be sampled. In a few cryo-EM 2D projections, ARMAN cells have a smoothly constricted cell wall (Figure 1b and Supplementary Figure 6d) suggestive of a division plane such as observed in Caulobacter crescentus (Comolli et al., 2005). Notably, dividing bacterial cells (Supplementary Figure 4) are 50–100 times more common than dividing ARMAN cells, consistent with a much slower ARMAN cell cycle.
Several cryogenic electron tomography 3D reconstructions of ARMAN revealed surface adhering lemon- (∼50 nm in diameter and ∼120–130 nm in length) and rod- (∼17–20 nm in diameter and very variable lengths reaching up to ∼300–350 nm) shaped objects. Based on their size, distinctive morphology (similar to spindle-shaped viruses, Prangishvili et al., 2006) and distribution, we infer that these are viruses (Figure 4, Supplementary Figure 6, and Supplementary movie 3). The large number of particles associated with individual cells (or in close proximity to them) suggest that cells have been imaged in the process of virus particle release, and that the release is non-lytic. The cell shown in Figure 4b has a total of 15 directly associated viral particles, the largest number we have counted. An image of larger field of view with a depth of 50-binned voxels, from transparent volume-rendered representation is provided in Supplementary Figure 6. Intriguingly, the findings may indicate near simultaneous replication of two very distinct viral types (superinfection). However, we cannot rule out the targeting of sensitive ARMAN cells by multiple viruses simultaneously. ARMAN cells associated with viruses exhibit a dramatic reorganization of the cytoplasm. In some cells with attached virus particles we observe almost no ribosomes (Supplementary Figure 6b). In most cases, the ribosomes loose their organization around the periphery of the cytoplasmic space (Figure 4b and Supplementary Figure 4f).
ARMAN and viruses. (a) A 50 voxel-thick slice through a volume-rendered tomographic reconstruction of an ARMAN cell surrounded by lemon- (segmented in yellow) and rod-shaped viruses (segmented in pink), some of which are fused with the cells and some of which are at a close distance but not attached to them (also see Supplementary Figure 6). Pili-like structures are shown in cyan. (b) A one voxel-thick slice through the 3D reconstruction of an ARMAN cell with many rod-like particles fused to it. Yellow arrows point to very high contrast elements in the cytoplasm (these are not present in uninfected cells). The top inset is the density plot profile along the blue line in (b) showing periodic mass density probably associated with the virus rods; the period is 9 pixels, or ∼11 nm. The bottom inset shows a lemon-shaped virus associated with the membrane of another ARMAN cell; the height of the inset box measures 134 nm.
Viral particles were commonly imaged in association with ARMAN cells, but this was not the case for other bacteria and archaea. The apparently relatively extensive targeting of ARMAN by viruses occurs despite their small cell volume and low ribosome copy number. The added metabolic cost of viral replication to the ARMAN cells may be substantial, and this may account for their typically low abundance in most AMD biofilms. In principle, to some extent the simultaneous challenges of low ribosome number and viral replication may be partially offset by an increase in cytoplasmic osmotic pressure as the result of small ARMAN cell volume, which should increase reaction rates and lower the copy number demand for protein complexes. The association of ribosomes along the inner membrane surface may enable highly orchestrated protein production, possibly also increasing translation efficiency. It could be also hypnotized that the ribosomes are in the periphery due to an exclusion caused by a nucleoid predominantly localized in towards the center; however, an advantage of some sort must result from such an organization, otherwise an alternative localization would have evolved. Finally, the ordering of complexes, possibly cytochromes, between the IM and OM likely optimizes space utilization in the periplasm and rates of material and energy transfer across the cell wall. A hypothesis that must be considered is that in combination, these features and possibly the tubular elements of unknown function, enable ARMAN cells to grow, albeit slowly, despite their small size and environmental challenges. A consequence of slow growth rates may be that isolation of these organisms will be difficult, making cultivation-independent deductions of their physiology and ecology all the more important.
References
Baker BJ, Banfield JF . (2003). Microbial communities in acid mine drainage. FEMS Microbial Ecol 44: 1–14.
Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Banfield JF . (2006). Lineages of acidophilic Archaea revealed by community genomic analysis. Science 314: 1933–1935.
Bond PL, Smigra SP, Banfield JF . (2000). Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an acid mine drainage site. Appl Environ Microbiol 66: 3842–3849.
Bremer H, Dennis PP . (1996). Modulation of chemical composition and other parameters of the cell growth rate in Escherichia coli and Salmonella: cellular and molecular biology. In: Neidhardt FC (ed). American Society for Microbiology, 2nd edn. 1553–1568 (http://ctbp.ucsd.edu/qbio/beemer96.pdf).
Comolli LR, Judd EM, Chen JC, Downing KH, Moerner WE, McAdams HH . (2005). Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol 187: 6874–6882.
Cox RA . (2004). Quantitative relationships for specific growth rates and macromolecular compositions of Mycobacterium tuberculosis, Streptomyces coelicolor A3(2) and Escherichia coli B/r: an integrative theoretical approach. Microbiol 150: 1413–1426.
Folk RL, Taylor LA . (2002). Nanobacterial alteration of pyroxenes in Martian meteorite Allan Hills 84001. Meteoritics & Planetary Science 37: 1057–1069.
Gilbert RJ, Fucini P, Connell S, Fuller SD, Nerhaus KH, Robinson CV et al. (2004). Three-dimensional structures of translating ribosomes by Cryo-EM. Mol Cell 14: 57–66.
Giovannoni SJ, Tripp JH, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M et al. (2006). Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103: 425–430.
Goldfarb DS . (2004). Microorganisms and calcium oxalate stone disease. Nephron Physiology 98: 48–54.
Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO . (2002). A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417: 63–67.
Hutchison III CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM et al. (1999). Global Transposon Mutagenesis and a Minimal Mycoplasma Genome. Science 286: 2165–2169.
Kajander EO, Çiftçioglu N . (1998). Nanobacteria: an alternative mechanism for pathogenic intra- and extracellular calcification and stone formation. Proc Natl Acad Sci USA 95: 8274–8279.
Kolisnychenko V, Plunkett III G, Herring CD, Fehér T, Pósfai J, Blattner FR et al. (2002). Engineering a reduced Escherichia coli genome. Genome Res 12: 640–647.
Kubitschek HE . (1990). Cell volume increase in Escherichia coli shifts to richer media. J Bacteriol 172: 94–101.
Lewin B . (1997). Genes VI. Oxford University Press: New York, ISBN 9780198577799.
Lewis PJ, Thaker SD, Errington J . (2000). Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J 19: 710–718.
Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL . (2004). Contribution of SAR11 bacteria to dissolved Dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl Environ Microbiol 70: 4129.
Marr AG . (1991). Growth rate of Escherichia coli. Microbiol Rev 55: 316–333.
Martel J, Young D-E . (2008). Purpoted nanobacteria in human blood as calcium carbonate nanoparticles. Proc Natl Acad Sci USA 105: 5549–5554.
Mascarenhas J, Weber MHW, Graumann PL . (2001). Specific polar localization of ribosomes in Bacillus subtilis. EMBO Rep 21: 685–689.
Miteva VI, Brenchley JE . (2005). Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland Glacier Ice Core. Appl Environ Microbiol 71: 7806–7818.
Nicastro D, Schwartz C, Pierson J, Cho J-C, Giovannoni SJ, McIntosh JR . (2006). Three-dimensional structure of the tiny bacterium Pelagibacter ubique studied by cryo-electron tomography. Microsc Microanal 12: 182–190.
Ortiz JO, Förster F, Kürner J, Linaroudis AA, Baumeister W . (2006). Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol 156: 334–341.
Prangishvili D, Forterre P, Garrett RA . (2006). Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4: 837–848.
Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake II RC et al. (2005). Community proteomics of a natural microbial biofilm. Science 308: 1915–1920.
Rappé MS, Connon SA, Vergin KL, Giovannoni SJ . (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633.
Sheridan PP, Miteva VI, Brenchley JE . (2003). Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland Glacier Ice Core. Appl Environ Microbiol 69: 2153–2160.
Steering Group for the Workshop on Size Limits of Very Small Microorganisms, National Research Council (1999). Size Limits of Very Small Microorganisms: Proceedings of a Workshop. National Academies Press: Washington, D.C., (http://www7.nationalacademies.org/ssb/nanomenu.html).
Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M et al. (2003). The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci 100: 12984–12988.
Acknowledgements
This research was supported by The Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy provided support under Contract No. DE-AC02-05CH11231 (LRC and KHD) and the US Department of Energy Genomics: GTL (JFB) program and by the NASA Astrobiology Institute (JFB). Mr Ted Arman (President, Iron Mountain Mines), Mr Rudy Carver, and Dr Richard Sugarek are thanked for site access and other assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is dedicated to the life and work of Professor Terry Beveridge.
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Comolli, L., Baker, B., Downing, K. et al. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J 3, 159–167 (2009). https://doi.org/10.1038/ismej.2008.99
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2008.99
Keywords
This article is cited by
-
Diversity, ecology and evolution of Archaea
Nature Microbiology (2020)
-
Perspectives on Cultivation Strategies of Archaea
Microbial Ecology (2020)
-
An archaeal symbiont-host association from the deep terrestrial subsurface
The ISME Journal (2019)
-
Eine oder zwei Membranen bei Prokaryoten: Was war zuerst da?
BIOspektrum (2019)
-
Cuniculiplasmataceae, their ecogenomic and metabolic patterns, and interactions with ‘ARMAN’
Extremophiles (2019)