Abstract
A fosmid clone 37F10 containing an archaeal 16S rRNA gene was screened out from a metagenomic library of Pearl River sediment, southern China. Sequence analysis of the 35 kb inserted fragment of 37F10 found that it contains a single 16S rRNA gene belonging to Miscellaneous Crenarchaeotal Group (MCG) and 36 open reading frames (ORFs). One ORF (orf11) encodes putative bacteriochlorophyll a synthase (bchG) gene. Bacteriochlorophyll a synthase gene has never been reported in a member of the domain Archaea, in accordance with the fact that no (bacterio)-chlorophyll has ever been detected in any cultivated archaea. The putative archaeal bchG (named as ar-bchG) was cloned and heterologously expressed in Escherichia coli. The protein was found to be capable of synthesizing bacteriochlorophyll a by esterification of bacteriochlorophyllide a with phytyl diphosphate or geranylgeranyl diphosphate. Furthermore, phylogenetic analysis clearly indicates that the ar-bchG diverges before the bacterial bchGs. Our results for the first time demonstrate that a key and functional enzyme for bacteriochlorophyll a biosynthesis does exist in Archaea.
Similar content being viewed by others
Introduction
Photosynthesis is arguably one of the most important biological processes on earth. However, the origin and evolution of photosynthesis still remain largely elusive, and controversial theories have been postulated (Meyer, 1994; Xiong et al., 2000; Xiong and Bauer, 2002b; Bryant and Frigaard, 2006a). Generally, photosynthesis is regarded as most likely having evolved after the divergence of the archaeal–eukaryal and bacterial lineages, as no (bacterio)-chlorophyll has ever been detected in a member of the domain Archaea. On the basis of genome comparisons, Raymond et al. (2002) postulated that horizontal gene transfer has played a major role in the evolution of bacterial phototrophs and that many of the essential components of photosynthesis have conducted horizontal gene transfer. Five phyla of bacteria including the cyanobacteria, proteobacteria (purple bacteria), green nonsulfur bacteria, green sulfur bacteria and the Gram-positive helicobacteria encompass the photosynthetic members. The purple bacteria and green nonsulfur bacteria synthesize a nonoxygen-evolving type II photosystem; the green sulfur bacteria and helicobacteria have a homodimeric type I photosystem; whereas cyanobacteria contain a type I photosystem and an oxygen-evolving type II photosystem, both of which are heterodimeric. The simple non-oxygen evolving photosystem is believed to be the ancester of the complex oxygen-evolving photosystem. These photosystems collect solar energy and convert it to chemical energy depending on photochemical reaction centers that contain chlorophylls or bacteriochlorophylls. These pigments are essential components of the photochemical reaction centers (Xiong and Bauer, 2002b; Bryant and Frigaard, 2006b).
Widespread in bacteria and ubiquitous in plants, chlorophylls and bacteriochlorophylls are involved fulfilling several functions in photosynthesis. The enzymes that involved in biosynthesis pathways of chlorophylls and bacteriochlorophylls have been largely identified and characterized. The chlorophyll biosynthesis is one of the intermediate steps in bacteriochlorophyll (Bchl) a biosynthesis; however, molecular phylogenetic analysis clearly indicates that Bchl a is a more ancient pigment (Willows, 2003). Biosynthesis of Bchl a needs esterifying isoprenoid tail by Bchl a synthase (BchG) from bacteriochlorophyllide a. BchG belongs to the UbiA prenyltransferase family of polyprenyltransferases with active motif DRXXD for binding of the divalent cations (Mg2+ or Mn2+) required for the catalytic activity (Lopez et al., 1996). The bchG genes in several photosynthetic bacteria have been identified by complementation of the mutated gene in vivo, or by heterologous expression and enzyme activity determination in vitro (Oster et al., 1997a). Until recently, bchG was only detected in photosynthetic organisms, therefore it has been utilized as a useful molecular marker for an evolutionary analysis of photosynthesis.
Recent progresses on genomic techniques have provided new opportunities to address challenging questions and to gain new perspectives on the microbial ecology and evolution (Venter et al., 2004; Green and Keller, 2006; Lasken, 2007; Martin-Cuadrado et al., 2007; Rusch et al., 2007). One of the most promising approaches, the metagenomic approach has been widely and successfully used in genome analysis of uncharacterized microbial taxa (Hallam et al., 2004, 2006; Moreira et al., 2004; Nunoura et al., 2005; Xu et al., 2007), expression of novel genes from uncultured environmental microorganisms (Schloss and Handelsman, 2003; Chung et al., 2008; Xu et al., 2008), elucidation of community-specific metabolism and comparison of gene contents in different communities (Culley et al., 2006; Green and Keller, 2006; Martin-Cuadrado et al., 2007). By using the metagenomic studies, our understanding of the bacterial and archaeal phototroph based on rhodopsin has been revolutionized (Frigaard et al., 2006; Walter et al., 2007). Until recently, prokaryotic rhodopsins were thought to exist exclusively in halophilic archaea. Metagenomic studies have revealed the existence, distribution and variability of a new class of such photoproteins, called proteorhodopsins, in members of the domain Bacteria (Beja et al., 2001; Venter et al., 2004). The easy lateral spread of rhodopsin throughout Archaea, Bacteria and Eucaryote were further discovered by the metagenomic studies (Frigaard et al., 2006). Metagenomic approach will facilitate a broader and deeper understanding of phototrophs, particularly in the community level (Bryant and Frigaard, 2006b). In this study, we report our discovery of a novel bacteriochlorophyll a synthase gene in an uncultivated archaea through the metagenomic approach. This is the first bchG found in a member of archaea.
Materials and methods
Metagenome sampling
Sediment was collected from an estuary station in Qi'ao Island (Pearl River Estuary, (E 113°38′07.3, N 22°27′21.4)) in Guangdong province, China, in April 2005 by using a single-core sampler. The length of the core is about 0.5 m, temperature of bottom water in this area was 21.5 °C and salinity concentration at the sediment surface was measured to be 2.6%. The sediments are soft silt, turned from gray on the surface layer to dark black only several cm below, accompanied with a light hydrogen sulfide smell. The core, which is 50 cm in length, was subsectioned into 2-cm slices and then transferred to sterile falcon tubes in a laminar flow cabinet and stored at −20 °C.
Fosmid library construction
The sediments from layer 16–32 cm were combined and used for fosmid library construction. The metagenomic library was constructed as follows: high molecular weight DNA was extracted according to the protocol described before (Xu et al., 2008), and loaded on pulsed field agarose gel electrophoresis after DNA ends were repaired by End-It DNA End-Repair Kit (Epicenter, Madison, WI, USA). After electrophoresis, an agarose plug containing 33–48 kb DNA was cut out. The genomic DNA purified from this plug was cloned into pCC1FOS (Epicenter). The ligated fosmids were packaged into MaxPlax Lambda Packaging Extract (Epicenter) and the packaged particles were transferred into Escherichia coli EPI300 (Epicenter). In total, nearly 8000 clones were obtained in this study and the average insert size was 35 kb.
Fosmid library screening and insert sequencing
PCR screening was conducted using the archaeal 16S rRNA gene-specific oligonucleotide primer set Arch21F and Arch958R (DeLong, 1992). PCR amplification involved 35 cycles of 95 °C 30 s, 55 °C 1 min, 72 °C 1 min and another step of 72 °C 10 min. The library was pooled into groups of 12 clones, which served for the screening. The fosmids were extracted by the standard alkaline lyses procedure from the pools of the library and used as templates for PCR. The fosmid pool, which was tested positive with the archaeal 16S rRNA gene-specific primers, was further screened by a PCR with each individual fosmid clone as template. The archaeal rRNA gene amplified from individual fosmid clones was sequenced using the Arch21F and Arch958R primers from both ends.
Fosmid clone sequence determination, annotation and confirmation
The fosmid clone sequence was determined by shotgun sequencing. Briefly, the plasmid was isolated and fragmented by sonication. Then, the fragmented DNA was separated by gel electrophoresis. Random 2 kb fragments were recovered from gels, blunt end-repaired and cloned into pUC18 vector at the SmaI site. The plasmids were sequenced from both ends using the ABI3700 sequencer (Applied Biosystem Inc., Foster City, CA, USA). The sequences generated had around 10-fold coverage of the inserted DNA. The sequences were assembled using the program Sequencer. Open Reading Frame (ORF) analysis was performed using the GeneMark Program (http://opal.biology.gatech.edu/GeneMark/). Translated amino acid sequences were used to search the GenBank, and EMBL databases with BLASTp (http://www.ncbi.nlm.nih.gov/BLAST/), and Wu-BLASTp (http://www.ebi.ac.uk/blast2/).
To make sure that the fosmid fragment does not represent artificial chimera during the cloning process, three pairs of primers targeting ORF17-19, ORF10-11 and ORF11-12 respectively were designed based on the fosmid DNA sequence to do PCR amplification from the environment DNA directly. The primer sequences are P1-F: TTTTTGGAGGGCGTTCTAAATGG; P1-R: ACTCCGCGGTTTTCGGGGTAGTT; P2-F: AATCATTGATAACAGCCAAAGTGTAGTA, P2-R: CTAGCTCCACATCAAAAACATTATTTAT; P3-F: CGTTGTTGTATTATGTTGCTTTGTCTGT, P3-R: TTTGGTTACTTCCTCCTTAGATGAGATG. The locations of the primers are illustrated on the genomic map of fosmid 37F10 in Figure 2. The PCR conditions used are the same as those for archaeal 16S rRNA gene amplification. The PCR products were extracted with a Gel-extraction kit (Omega Bio-Tek Inc., Norcross, GA, USA). Afterward, the purified DNA products were ligated with the pMD18-T vector (Takara, Dalian, Liaoning Province, China) and transformed to competent cells of E. coli DH-5α according to the manufacturer's instructions. Three positive clones from each PCR product were sequenced.
Heterologous expression of ar-bchG in E.coli
The ar-bchG expression plasmid was constructed by polymerase chain reaction (PCR) amplification of 37F10 plasmid. The forward primer (5′-CCGGTGCATGCATATGTTTAGTAGTTTGAGCGGTT-3′) was designed to contain an SphI restriction site (underlined) introduced at the translation start site. The reverse primer (5′-CCGGTAGATCTGAATAACACATTAGGTATTTTC-3′) was designed to contain a BglII restriction site upstream the translation stop site. PCR amplification involved 30 cycles of 95 °C 30 s, 55 °C 1 min, 72 °C 1 min and another step of 72 °C 10 min. The PCR-amplified ar-bchG gene was purified by agarose gel electrophoresis and cloned into the expression vector pQE70 (Qiagen, Hilden, Germany) following the manufacturer's instructions. The plasmid pAr-bchG was transformed into E. coli strain M15 and over expressed following the manufacturer's instructions.
Preparation of pigments and Bacteriochlorophyll synthase assays
Bacteriochlorophyllide a was prepared as described before (Fiedor et al., 1992; Oster et al., 1997b) using leaves of Ailanthus altissima as the source of chlorophylase.
Bacteriochlorophyll synthase assay was carried out according to Oster et al. (1997a) with some modification. Aliquots of this bacterial lysate containing ∼20 mg of protein were diluted with 200 μl of reaction buffer (120 mM potassium acetate, 10 mM magnesium acetate, 50 mM Hepes/KOH, pH 7.6, 14 mM mercaptoethanol and 10% glycerol), 30 μl of 5 mM ATP, and 10 μl of 4 mM geranylgeranyl diphosphate or phytyl diphosphate. The reaction was then started by addition of 10 μl of 0.1 mM bacteriochlorophyllide a. The other reaction procedures and analytic HPLC were performed according to Oster et al. (1997a).
Phylogenetic analysis
The archaeal 16S rRNA gene phylogenetic tree was constructed using Mega 4.0 based on neighbor-joining method with 1000 bootstrap. Amino acid sequences of some typical enzymes classified as members of the ‘UbiA prenyltransferase family’ according to the Pfam protein family database (www.sanger.ac.uk/software/Pfam/) were obtained from public protein databases. They were aligned using the ClustalX 1.83 program and the phylogenetic tree was constructed with the maximum likelihood method based on Jones–Taylor–Thornton model by Phylip 3.67 package. 1000 trial of bootstrap analysis was used for calculation.
Accession number
The sequence of fosmid 37F10 has been submitted to GeneBank, the accession number is EU559699.
Results and discussion
Fosmid library construction and screening
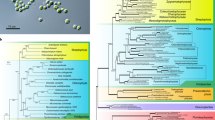
A microbial diversity investigation of a sediment core near Qi'ao Island in the Pearl River of southern China revealed a unique microbial community with a large number of uncharacterized archaea, and the middle layers of the sediment core exhibited higher archaeal diversity (unpublished observations, LJ Jiang). To obtain more genetic information, and to infer the physiology of these archaea, a fosmid library was constructed from middle layers of the sediment core (16–32 cm). More than 8000 clones were obtained in the fosmid library with average insert length of around 35 kbp (data not shown). The fosmid library was screened by PCR amplification with archaeal 16S rRNA gene primers (arch21F/958R). Three fosmid clones containing archaeal 16S rRNA gene was screened out, and one fosmid clone named 37F10 containing a 16S rRNA gene, which belongs to the Miscellaneous Crenarchaeotal Group (MCG) was sequenced. The Miscellaneous Crenarchaeotal Group distributed from the top to the bottom layer along the sediment core (our unpublished data, LJ Jiang). The 16S rRNA gene on clone 37F10 had highest identity (95%) with a sequence isolated from solid waste landfill (Huang et al., 2005). Phylogenetic analysis showed that the fosmid-derived archaeal 16S rRNA gene could be assigned into MCG (Inagaki et al., 2003) (Figure 1). The MCG archaea are found globally distributed in both surface and subsurface environments, indicating a high ecophysiological flexibility (Biddle et al., 2006; Sorensen and Teske, 2006). To date, no cultivated MCG archaea are available and almost no metabolic or physiological properties of this group of archaea are known, except that it was suggested to have a heterotrophic lifestyle based on stable isotope analysis (Biddle et al., 2006).
Phylogenetic tree based on archaeal 16S rRNA genes. The 1.5 kb nucleotide positions of the 16S rRNA genes were aligned with clustalX 1.83 program. The phylogenetic tree was constructed from a matrix by least-squares distance matrix analysis (Olsen, 1988) and the neighbor-joining method (Saitou and Nei, 1987) using Mega 4. The Euryarchaeota Methanococcus vannielii and Thermoplasma acidophilum were used as outgroups. The reference sequences from MCG were chosen as they show high sequence identity with 16S rRNA gene on the fosmid clone 37F10. Sequences from MBGB and MGI selected here are those frequently cited reference sequences in these two groups. A 1000 trial of bootstrap analysis was used to provide confident estimates for phylogenetic tree topology. The scale bar represents number of substitutions per site. 37F10 refers to the 16S rRNA gene on the fosmid clone 37F10; MCG: Miscelleous Crenarchaeota Group (Takai et al., 2001; Inagaki et al., 2003) ; MBGB: Marine Benthic Group B (Vetriani et al., 1999) and MGI: Marine Group I (DeLong, 1992; Fuhrman et al., 1992)
Characterization of genome fragment from MCG
The fosmid clone 37F10 was fully sequenced and found to contain a 35 kb insert sequence with 52.48% G+C content that contained 36 predicted open reading frames (ORFs) plus a single 16S rRNA gene (Table1 and Figure 2). Normally, most of the known Archaea have one or a few copies of rRNA operon containing at least both 16S and 23S rRNA gene (Nunoura et al., 2005). However, the separated localization of 16S and 23S rRNA gene on the genome has also been identified in Nanoarchaeota: Nanoarchaeum equitans (Waters et al., 2003), and several Euryarchaea (Ruepp et al., 2000; Beja et al., 2001; Slesarev et al., 2002). In addition, Nunoura et al. has also reported the finding of a fosmid clone which contained a single 16S rRNA gene from hot water crenarchaeotic group I of Crenarchaeota (Nunoura et al., 2005).
Genomic map of fosmid clone 37F10. The complete DNA sequence of the clone was determined by using the short-gun sequencing method and the GeneMark Program was used to perform Open Reading Frame (ORF) analysis. Clone 37F10 is 34528 bp long and contains 36 putative ORF plus a 16S rRNA gene. The number of the ORF is given on top of the gene. The locations of the primers to amplify fragment P1, P2, P3 targeting ORF17-19, ORF10-11, ORF11-12, respectively, were presented on top of the sequence. The putative origins of the genes (bacteria, archaea or euryarchaea) were illustrated by different colors of the arrows. The putative functions of the encoded proteins are also given when homology search gave e-values less than 1E-04 and HP indicates ‘the hypothetic protein’.
Sequence blast analysis demonstrated that the majority of the predicted proteins encoded by ORFs upstream and downstream of the 16S rRNA gene (ORF1 to ORF17) had their highest similarity to archaeal homologs (Table 1). Meanwhile, most of the predicted proteins encoded by ORFs downstream ORF 17 (ORF18 to ORF36) had the highest similarity to homologs of bacterial origin. The mean G-C% value in the ‘archaeal like half’ (ORF1-17) was 42.4%, whereas that was 60.1% in the ‘bacterial like half’ (ORF18-36). The harboring of genes in a genome fragment from both archaeal and bacterial origin has frequently been observed, which suggest extensive horizontal gene transfer between archaea and bacteria (Nelson et al., 1999; Deppenmeier et al., 2002). The genomic sequence in the fosmid 37F10 has very likely conducted horizontal gene transfer. To prove that the genome fragment cloned in the fosmid was not an artificial chimera generated in the cloning process, three sets of primers were designed to amplify DNA fragments P1, P2 and P3 encompassing ORFs of greatest interests or concerns in this study from sediment (see Figure 2 for primer and DNA fragments' location). Specific PCR bands could be successfully obtained from the sediment DNA using all the three sets of primers (Supplementary Figure S1). The sequences of the PCR products from the sediment were determined and showed to be the same as those from the fosmid 37F10. The successful amplification of fragment P1, P2 and P3 from sediment DNA clearly indicates that the DNA fragment in the fosmid represents the original DNA fragment from the sediment.
Surprisingly, an ORF (ORF11) encoding a putative bacteriochlorophyll a synthase (BchG) was found locating closely to the 16S rRNA gene. The inferred amino acid sequence of the putative bacteriochlorophyll a synthase showed high identity (27% aa identity, E value=le-06) with BchG from the photosynthetic bacterium Rhodospirillum rubrum. All of the ORFs surrounding the putative bchG gene (named as ar-bchG) encoded putative proteins with their highest similarity to proteins from archaea, except one of eukaryotic origin (Table 1, Figure 2). The ar-bchG gene encoded a polypeptide of 299 amino acids, with molecular weight of 34 kDa. Hydropathy plots indicated seven transmembrane domains and a signal peptide fragment, a typical feature of the UbiA prenyltransferase family. Alignment of amino acid sequences of members of UbiA prenyltransferase superfamily clearly indicates the presence of a conserved domain, which contains the DRXXD motif (Supplementary Figure S2). The DRXXD motif is proposed to be responsible for the binding of the divalent cations (Mg2+ or Mn2+) required for the catalytic activities of polyprenyltransferases (Lopez et al., 1996). We found that the Arginine in the DRXXD motif is not much conserved even in the members of ChlG/BchG subcluster, it could be substituted by other amino acids such as Valine, Alanine or Leucine (Supplementary Figure S2).
Phylogenetic analysis of UbiA prenyltransferase family proteins
Bacteriochlorophyll/chlorophyll synthetase is a subfamily (ChlG/BchG subfamily) of UbiA prenyltransferase family (a large family of polyprenyltransferases) which contains several distinct enzyme clusters and each of the enzyme clusters accepts specific prenyl-acceptors with similar structures (Hemmi et al., 2004b). Members of ChlG/BchG subfamily are found exclusively in photosynthetic organisms; therefore it has been used as a useful biomarker for detection and evolutionary analysis of photosynthesis.
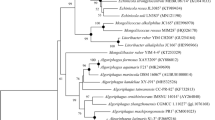
Amino acid sequences of some typical enzymes classified as members of the ‘UbiA prenyltransferase family’ according to the Pfam protein family database were obtained and the phylogenetic tree was constructed with the maximum likelihood method as shown in Figure 3. The archaeal BchG (Ar-BchG) clustered with the bacterial BchGs, as shown in the phylogenetic tree of the UbiA prenyltransferases (Figure 3), forming the BchG/ChlG subgroup, separated from any other UbiA prenyltransferase clusters. Moreover, the phylogenetic analysis clearly indicates that Ar-BchG forms a distinct branch from the known photosynthetic bacterial BchGs and it diverges earlier than photosynthetic bacterial BchGs.
Phylogenetic analysis of UbiA prenyltransferase family proteins. The phylogenetic relationship of the UbiA prenyltransferases from the six clusters of subfamilies, that is, ChlG/BchG (represented by green color), UbiA/COQ2 (orange), CyoE/COX10 (pink), DGGGPS (light blue), MenA (yellow), HPT(purple) were determined using the maximum-likelihood method with phylip package 3.67. Abbreviations and accession numbers of each protein are as follows: 37F10, putative Ar-BchG from fosmid 37F10; ChcBchG, bacteriochlorophyll synthase subunit BchG from Chlorobium chlorochromatii CaD3 (YP_378530); SynChlG, chlorophyll a synthase ChlG from Synechococcus sp. WH 8102 (NP_897768); ChaBchG, bacteriochlorophyll synthase BchG from Chloroflexus aurantiacus J-10-fl (ZP_ 00767878); HmBchG, bacteriochlorophyll synthase subunit BchG from Heliobacillus mobilis (AAC84024); RcBchG, bacteriochlorophyll synthase subunit BchG from Rhodobacter capsulatus (CAA77532); R.rubrum BchG, bacteriochlorophyll synthase subunit BchG from Rhodospirillum rubrum ATCC 11170 (YP_425718); SynHPT, homogentisate phytyltransferase Slr1736 from Synechocystis sp. strain PCC 6803 (BAA17774); AtHPT1, homogentisate phytyltransferase HPT1 from Arabidopsis thaliana (AAM10489); BcMenA, 1,4-dihydroxy 2-naphtoate polyprenyltransferase MenA from Bacillus cereus (AAP11757); EcMenA, 1,4-dihydroxy 2-naphtoate octaprenyltransferase MenA from E. coli (AAC76912); APE0159, probable (S)-2,3-Di-O-farnesylgeranylglyceryl synthase (BAA79070); HalHhoA, HhoA protein from Halobacterium sp. NRC-1, annotated as the 4-hydroxybenzoate octaprenyltransferase (AAG19118); TA0996, a hypothetical protein from Thermoplasma acidophilum, annotated as the predicted 4-hydroxybenzoate polyprenyltransferase (NP_394456); MjUbiA, UbiA protein from Methanocaldococcus jannaschii, annotated as the 4-hydroxybenzoate octaprenyltransferase (AAB98267); SsUbiA2, DGGGPS from Sulfolobus solfataricus named UbiA-2 (AAK40896); PH0027, a hypothetical protein from Pyrococcus horikoshii OT3 (BAA29095); SsUbiA1, UbiA-1 protein from S. solfataricus, annotated as the 4-hydroxybenzoate octaprenyltransferase (AAK40480); EcUbiA, 4-hydroxybenzoate octaprenyltransferase UbiA from E. coli (AAC43134); LePGT1, 4-hydroxybenzoate geranyltransferase PGT-1 from Lithospermum erythrorhizon (BAB84122); ScCOQ2, 4-hydroxybenzoate hexaprenyltransferase COQ2 from S. cerevisiae (CAA96321); SSO0656, hypothetical protein from S. solfataricus, annotated as the cytochrome c oxidase folding protein (AAK40961); EcCyoE, protoheme IX farnesyltransferase CyoE from E. coli (AAC73531); and ScCOX10, protoheme IX farnesyltransferase COX10 from Saccharomyces cerevisiae (CAA97879).
Although the phylogenetic linkage between Ar-BchG and the members of ChlG/BchG looks week (Figure 3, low bootstrap value), however, the close relationship and consistency of the tree topology grouping Ar-BchG and members of ChlG/BchG together have been provided by our phylogenetic analysis using two different methods including Maximum-likelihood and Neighbor-joining (Figure3 and Supplementary Figure S3). As Ar-BchG showed closer relationship with enzymes from members of ChlG/BchG family, and didn't show any relationship with members of other known subclusters in UbiA superfamily, it was temporally placed into the ChlG/BchG subcluster here. However, it should also be noticed that Ar-BchG forms a distinct branch from known photosynthetic bacterial BchGs, it is possible that Ar-BchG may form a new subcluster if more related sequences could be obtained later. We searched in the public databases for other archaeal sequences related with Ar-BchG, none was found to cluster with Ar-BchG (data not shown). Previously, protein APE0159 from the marine aerobic hyperthermophilic crenarcheon Aeropyrum pernix K1(BAA79070) was annotated as a putative bacteriochlorophyll synthase; however, it was later found by phylogenetic analysis that the protein belongs to DGGPS subcluster (Hemmi et al., 2004b), therefore, it is currently annotated as ‘probable (S)-2,3-Di-O-farnesylgeranylglyceryl synthase’ in databank.
Cloning and heterologous expression of ar-bchG
Hemmi et al. (2004b) have suggested that the position of the prenyltransferase in the UbiA protein phylogenetic tree can be used to infer its specificity for a prenyl-acceptor substrate. BchG catalyzes the esterification of bacteriochlorophyllide a with phytol or geranylgeraniol (Garcia-Gil et al., 2003; Willows, 2003). The clustering of Ar-BchG with BchGs in the phylogenetic tree leads us to suspect that Ar-BchG may function in the synthesis of bacteriochlorophyll a from bacteriochlorophyllide a and phytyl diphosphate or geranylgeranyl diphosphate (GGPP).
To determine whether Ar-BchG synthesize bacteriochlorophyll a from bacteriochlorophyllide a and phytyl diphosphate or geranylgeranyl diphosphate (GGPP). The ar-bchG gene was PCR amplified and cloned into the expression vector pQE70. The formed expression plasmid pAr-bchG was transformed into E. coli M15. The membrane proteins extracted from the E.coli strain containing the expression plasmid pAr-bchG were used to check its enzyme activity. High performance liquid chromatography (HPLC) with fluorescence detection was used to differentiate the esterified bacteriochlorophyllide a from its substrate (Hemmi et al., 2004a). Bacteriochlorophyll a from Rhodoseudomonas sphaeroides purchased from Sigma was used here as a standard. As shown in Figure 4, the expressed protein was capable of synthesizing bacteriochlorophyll a using bacteriochlorophyllide a and GGPP or Phytyl-PP as substrates. No esterification was found when the same substrates were incubated with extracts from the non-transformed E. coli strain (data not shown). These results clearly demonstrate that the putative bchG of crenarchaeota indeed has bacteriochlorophyll synthase activity. This is the first functional bacteriochlorophyll synthase ever found originating from Archaea.
HPLC elution profile of bacteriochlorophyll a synthesized by the heterologously overexpressed bchG gene product. The results of incubating E.coli bchG expression extracts with bacteriochlorophyllide a and phytyl diphosphate (a) or geranylgeranyl diphosphate (b). Peak 1 bacteriochlorophyll a esterified with phytol-PP (the majority of non-esterified pigment was removed prior to HPLC by phase separation with n-hexane); peak 2 is bacteriochlorophyll a esterified with geranylgeranyl-PP (GGPP).
The UbiA superfamily enzymes catalyze the transfer of a prenyl (or phytyl) group to hydrophobic acceptors whose structures vary extensively. The subclusters in the UbiA superfamily recognize prenyl-acceptors of similar structures, and are involved in the biosynthesis of various substances, for instance, quinines (UbiA/COQ2 subcluster), hemes (CyoE/COX10), chlorophylls (ChlG/BchG), membrane lipids (DGGGPS). High substrate specificity has been observed for the enzymes. Ar-BchG has the function to esterify bacteriochlorophyllide a; however, we could not rule out the possibility that Ar-BchG may probably utilize other substrates with unknown structure to us at present. Nevertheless, our study at least indicates for the first time that an archaeal-derived enzyme of UbiA family has the function of BchG which has never been thought possible before.
Implication for photosynthesis evolution
In photosynthetic bacteria, the bchG gene is found either clustered with other photosynthetic genes or as a single gene alone in the genome without forming any clusters (Xiong and Bauer, 2002a). On the fosmid clone 37F10, only the ar-bchG gene, which is involved in bacteriochlorophyll biosynthesis, was identified; no other bch genes could be clearly interpreted through BLAST search on the 35 kb insert sequence (except that a geranylgeranyl diphosphate synthase gene was found adjacent to the ar-bchG gene on the fosmid clone). However, the verification of the function of ar-bchG suggests that the uncultivated crenarchaeota may have the potential to synthesize bacteriochloropyll a. If this is true, what is the ecological function or benefit for having the ability to synthesize bacteriochlorophyll in MCG, which reside predominantly in sediments or soils? MCG sequences were first found from the deep terrestrial subsurface in South African goldmines, they form ‘Terrestrial Miscellaneous Crenarchaeotic Group’ with sequences from other terrestrial habitats (Takai et al., 2001). Later, it was found that MCG sequences are not restricted within terrestrial habitats, and renamed as ‘Miscellaneous Crenarchaeotic Group’ (Inagaki et al., 2003). At present, nearly nothing is known about MCG except that it was found cosmopolitan in various environments including petroleum-contaminated soil (Kasai et al., 2005), estuarine sediment (this study and our unpublished data), marine sediments (Inagaki et al., 2006), subsurface thermal spring (Weidler et al., 2007), shallow submarine hot spring (Hirayama et al., 2007) and hydrothermal vent sediments (Nercessian et al., 2005). In our case, although the archaeal fosmid clone containing ar-bchG was isolated from sediment layer of 16–32 cm of the sediment core, which should be a typical dark environment, MCG was found distributing from the surface to the bottom of the core (our unpublished data). We suppose that containing a presumptive Bchl a synthase gene may give the archaea more flexibility to survive or adapt to various environments.
To date, bacteriochlorophyll biosynthesis has never been detected in any archaeal organism. Thus, the origin of photosynthesis is believed to have occurred after the divergence of Archaea and Eubacteria. The discovery of a functional enzyme involved in Bchl biosynthesis in Archaea reported here raises the significant possibility that the origin of photosynthesis probably predates the divergence of bacteria and archaea. On the other hand, one should be aware that the identification of a gene-encoding protein with functional BchG activity does not mean that is what it does in vivo. It would be possible that Ar-BchG has several functions other than bacteriochlorophyll a synthesis, such as synthesis of membrane lipids. Nevertheless, the finding of a protein having BchG activity (in vitro) from archaea should at least let us reconsider the evolution of this protein family. Archaea should have played an important role in the molecular evolution of (bacterio)-chlorophyll a synthase, which has never been found before.
Accession codes
References
Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF . (2001). Proteorhodopsin phototrophy in the ocean. Nature 411: 786–789.
Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sorensen KB, Anderson R et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103: 3846–3851.
Bryant DA, Frigaard NU . (2006a). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14: 488–496.
Bryant DA, Frigaard NU . (2006b). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14: 488–496.
Chung EJ, Lim HK, Kim JC, Choi GJ, Park EJ, Lee MH et al. (2008). Forest soil metagenome gene cluster involved in antifungal activity expression in Escherichia coli. Appl Environ Microbiol 74: 723–730.
Culley AI, Lang AS, Suttle CA . (2006). Metagenomic analysis of coastal RNA virus communities. Science 312: 1795–1798.
DeLong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R et al. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4: 453–461.
Fiedor L, Rosenbach-Belkin V, Scherz A . (1992). The stereospecific interaction between chlorophylls and chlorophyllase. Possible implication for chlorophyll biosynthesis and degradation. J Biol Chem 267: 22043–22047.
Frigaard NU, Martinez A, Mincer TJ, DeLong EF . (2006). Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439: 847–850.
Fuhrman JA, McCallum K, Davis AA . (1992). Novel major archaebacterial group from marine plankton. Nature 356: 148–149.
Garcia-Gil LJ, Gich FB, Fuentes-Garcia X . (2003). A comparative study of bchG from green photosynthetic bacteria. Arch Microbiol 179: 108–115.
Green BD, Keller M . (2006). Capturing the uncultivated majority. Curr Opin Biotechnol 17: 236–240.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95.
Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM et al. (2004). Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science 305: 1457–1462.
Hemmi H, Shibuya K, Takahashi Y, Nakayama T, Nishino T . (2004a). (S)-2, 3-Di-O-geranylgeranylglyceryl phosphate synthase from the thermoacidophilic archaeon Sulfolobus solfataricus. Molecular cloning and characterization of a membrane-intrinsic prenyltransferase involved in the biosynthesis of archaeal ether-linked membrane lipids. J Biol Chem 279: 50197–50203.
Hemmi H, Shibuya K, Takahashi Y, Nakayama T, Nishino T . (2004b). (S)-2, 3-Di-O-geranylgeranylglyceryl phosphate synthase from the thermoacidophilic archaeon Sulfolobus solfataricus. Molecular cloning and characterization of a membrane-intrinsic prenyltransferase involved in the biosynthesis of archaeal ether-linked membrane lipids. J Biol Chem 279: 50197–50203.
Hirayama H, Sunamura M, Takai K, Nunoura T, Noguchi T, Oida H et al. (2007). Culture-dependent and -independent characterization of microbial communities associated with a shallow submarine hydrothermal system occurring within a coral reef off Taketomi Island, Japan. Appl Environ Microbiol 73: 7642–7656.
Huang LN, Zhu S, Zhou H, Qu LH . (2005). Molecular phylogenetic diversity of bacteria associated with the leachate of a closed municipal solid waste landfill. FEMS Microbiol Lett 242: 297–303.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments, on the Pacific Ocean Margin. P Natl Acad Sci USA 103: 2815–2820.
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of okhotsk. Appl Environ Microbiol 69: 7224–7235.
Kasai Y, Takahata Y, Hoaki T, Watanabe K . (2005). Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ Microbiol 7: 806–818.
Lasken RS . (2007). Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol 10: 510–516.
Lopez JC, Ryan S, Blankenship RE . (1996). Sequence of the bchG gene from Chloroflexus aurantiacus: relationship between chlorophyll synthase and other polyprenyltransferases. J Bacteriol 178: 3369–3373.
Martin-Cuadrado AB, Lopez-Garcia P, Alba JC, Moreira D, Monticelli L, Strittmatter A et al. (2007). Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE 2: e914.
Meyer TE . (1994). Evolution of photosynthetic reaction centers and light harvesting chlorophyll proteins. Biosystems 33: 167–175.
Moreira D, Rodriguez-Valera F, Lopez-Garcia P . (2004). Analysis of a genome fragment of a deep-sea uncultivated Group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ Microbiol 6: 959–969.
Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH et al. (1999). Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399: 323–329.
Nercessian O, Fouquet Y, Pierre C, Prieur D, Jeanthon C . (2005). Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ Microbiol 7: 698–714.
Nunoura T, Hirayama H, Takami H, Oida H, Nishi S, Shimamura S et al. (2005). Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ Microbiol 7: 1967–1984.
Olsen GJ . (1988). Phylogenetic analysis using ribosomal RNA. Methods Enzymol 164: 793–812.
Oster U, Bauer CE, Rudiger W . (1997a). Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Biol Chem 272: 9671–9676.
Oster U, Bauer CE, Rudiger W . (1997b). Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Biol Chem 272: 9671–9676.
Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE . (2002). Whole-genome analysis of photosynthetic prokaryotes. Science 298: 1616–1620.
Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW et al. (2000). The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407: 508–513.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77.
Saitou N, Nei M . (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Schloss PD, Handelsman J . (2003). Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14: 303–310.
Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV et al. (2002). The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci USA 99: 4644–4649.
Sorensen KB, Teske A . (2006). Stratified communities of active Archaea in deep marine subsurface sediments. Appl Environ Microbiol 72: 4596–4603.
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK . (2001). Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67: 5750–5760.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL . (1999). Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol 65: 4375–4384.
Walter JM, Greenfield D, Bustamante C, Liphardt J . (2007). Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA 104: 2408–2412.
Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M et al. (2003). The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA 100: 12984–12988.
Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H . (2007). Communities of Archaea and Bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl Environ Microb 73: 259–270.
Willows RD . (2003). Biosynthesis of chlorophylls from protoporphyrin IX. Nat Prod Rep 20: 327–341.
Xiong J, Bauer CE . (2002a). Complex evolution of photosynthesis. Annu Rev Plant Biol 53: 503–521.
Xiong J, Bauer CE . (2002b). Complex evolution of photosynthesis. Annu Rev Plant Biol 53: 503–521.
Xiong J, Fischer WM, Inoue K, Nakahara M, Bauer CE . (2000). Molecular evidence for the early evolution of photosynthesis. Science 289: 1724–1730.
Xu M, Wang F, Meng J, Xiao X . (2007). Construction and preliminary analysis of a metagenomic library from a deep-sea sediment of east Pacific Nodule Province. FEMS Microbiol Ecol 62: 233–241.
Xu M, Xiao X, Wang F . (2008). Isolation and characterization of alkane hydroxylases from a metagenomic library of Pacific deep-sea sediment. Extremophiles 12: 255–262.
Acknowledgements
We are very grateful to Professors Carl E Bauer (Indiana University), Robert E Blankenship (Arizona State University), Donald A Bryant (The Pennsylvania State University), and Robert D Willows (Macquarie University, Australia) for critical reading of the paper and useful suggestions for the work. We thank Mr Ke Xu, Dr Zhen Wu and Dr Weihong Li (Xiamen University) for their help in HPLC analysis. This work is partly supported by NSFC grant (40625016, 40532011), China Ocean Mineral Resources R&D Association (COMRA DYLY0202-01).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Meng, J., Wang, F., Wang, F. et al. An uncultivated crenarchaeota contains functional bacteriochlorophyll a synthase. ISME J 3, 106–116 (2009). https://doi.org/10.1038/ismej.2008.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2008.85
Keywords
This article is cited by
-
Genomic and transcriptomic dissection of Theionarchaea in marine ecosystem
Science China Life Sciences (2022)
-
Diversity, ecology and evolution of Archaea
Nature Microbiology (2020)
-
A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments
Microbiome (2018)
-
Distribution of Bathyarchaeota Communities Across Different Terrestrial Settings and Their Potential Ecological Functions
Scientific Reports (2017)
-
Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage
The ISME Journal (2016)