Abstract
A sequence analysis is described of a fosmid clone from a coastal marine metagenomic library that contains a 16S rRNA gene with high sequence similarity to that of the SAR11 bacterium ‘Candidatus Pelagibacter ubique’ HTCC1062. The sequence of the fosmid clone was 32 086 bp in length and contained 23 187 bp of the 48-kb hyper-variable region 2 (HVR2) present in the genome of ‘Cand. P. ubique’. However, half of the sequences within the HVR2 region of the fosmid clone show little sequence similarity to or have no representative homologues in the genome sequence of ‘Cand. P. ubique’ HTCC1062. Given their putative functions, the acquisition of these genes suggests that SAR11 could harbour more diverse phenotypes than represented by the 16S rRNA taxonomy. Variation in SAR11 genomes from different locations might explain why SAR11 is abundant in so many diverse marine provinces.
Similar content being viewed by others
Main
Recent analysis of the Sargasso Sea and Global Ocean Survey metagenomes has provided evidence of regions of hyper variability in the genomes of SAR11 (Rusch et al., 2007; Wilhelm et al., 2007). This was achieved using fragment recruitment (Wilhelm et al., 2007), whereby environmental sequence data were compared against the genome of ‘Candidatus Pelagibacter ubique’ HTCC1062 and then assembled (Rusch et al., 2007).
We have confirmed a region of hyper variability in an SAR11 genome by screening a metagenomic fosmid library for SAR11-like 16S rRNA gene-containing clones. SAR11-specific 16S rRNA gene primers (Béjà et al., 2000) were used to identify clones that cover the 48-kb hyper-variable region 2 (HVR2) of ‘Cand. P. ubique’ HTCC1062. A clone was identified that contained an SAR11 16S rRNA gene sequence from a library of 10 000 clones, constructed with surface seawater from the Western Channel Observatory (WCO: http://www.westernchannelobservatory.org.uk/, 50.25°N: 04.212°W) (see Supplementary Information for description of methods).
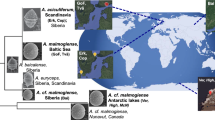
PCR screening of the fosmid library yielded only one positive clone (clone 01–003783: GenBank accession number EU410957; G/C content 43.2%), which contained a 16S rRNA gene with 98.8% sequence identity to that of ‘Cand. P. ubique’ HTCC1062. Phylogenetic analysis of the 16S rRNA gene sequence confirmed that it belongs to the SAR11 clade of bacteria (Supplementary Figure S1). The small number of SAR11 fosmid clones produced is consistent with the findings of others (Suzuki et al., 2004; DeLong et al., 2006) and is puzzling given the high abundance and ubiquitous distribution of this clade (Morris et al., 2002). It has been suggested that underrepresentation might be due to the presence of genes, in proximity to the ribosomal RNA operon, which express products toxic to the host cell (Béjà et al., 2000). To our knowledge, there is no evidence of such genes from published genomes.
Table 1 summarizes the identity and location of each coding sequence (CDS) of fosmid clone 01–003783. Apart from the ribosomal RNA operon, a total of 25 complete and one partial CDS were predicted. Of these, only 16 showed high sequence similarity to genes from ‘Cand. P. ubique’ HTCC1062 (Table 1). To exclude the possibility that inclusion of these CDSs in the fosmids resulted from a chimeric cloning event, PCR primers were designed to amplify fragments between each of the CDSs from 13 to the rRNA operon (Figure 1). All of these regions were detected in the environmental DNA used to produce the fosmid library. As all CDSs were complete, a chimera could only have formed between the CDSs. By amplifying and sequencing these regions from environmental DNA, we have demonstrated that this pattern of genes existed prior to library construction and hence cannot be a chimera.
Diagram of the fosmid clone sequence. Large arrows represent relative size and orientation of open reading frames (ORFs). 16S, 23S rRNA and the intergenic transcribed spacer region (ITS) are represented by two boxes titled 23S and 16S. Small solid arrows represent regions amplified by PCR from environmental DNA to validate clone structure. Core conserved genes represent area with synteny to ‘Candidatus Pelagibacter ubique’.
Only the four CDSs transcribed in the 3′-direction after the rRNA operon shared synteny with the ‘Cand. P. ubique’ HTCC1062 and HTCC1002 genomes (Table 1). The 11 CDSs with no homologue in the genome of HTCC1062 represent potential niche-specific acquisitions that may provide a competitive advantage (Wilhelm et al., 2007). For example, acquisition of a gene involved in 5′-nucleotidase and uridine diphosphate (UDP)-sugar hydrolysis (CDS 5) might benefit utilization of extracellular nucleotides as a phosphate source under conditions of phosphate starvation (Rittmann et al., 2005). The missing gene products required for these processes to be fully functional may be encoded within the 25 kb of the 48-kb large HVR2 that were not part of the sequence of fosmid clone 01–003783. However, we were not successful in identifying a contiguous clone within the fosmid clone library. Their absence suggests that these genes are either non-functioning or involved in other processes. The former is unlikely due to the genomic streamlining of SAR11 organisms (Giovannoni et al., 2005), which would result in the loss of non-functioning elements.
It is interesting that 22 of the CDSs of fosmid clone 01–003783 show higher sequence similarity and have a closer phylogenetic relationship to the respective sequences of bacteria not belonging to the SAR11 clade (Table 1; Supplementary Figure S2). This is in contrast to the phylogenetic inference based on the 16S rRNA gene sequence within the same fosmid clone (Supplementary Figure S1). A possible explanation for this may be that CDSs 1–22 are a result of horizontal gene transfer. Although there is no direct evidence for phage-mediated horizontal gene transfer in fosmid clone 01–003783, the possibility is supported by the low G/C content (potentially indicative of phage genes) of six of the CDSs in the fosmid (Table 1). Also, phage integrase genes have been documented in the other 49% of HVR2 that was not in this fosmid clone (Rusch et al., 2007). The ecological ramifications of horizontal gene transfer for SAR11 bacteria have been discussed previously (for example, Rusch et al., 2007; Wilhelm et al., 2007).
In conclusion, the presence of a region of extreme variability within bacterial genomes (Rusch et al., 2007, Wilhelm et al., 2007) has been confirmed in fosmid clone 01–003783; this is a rare example of a fosmid that contains the SAR11 16S rRNA gene. The variable region HVR2 provides evidence in support of the hypothesis of bacterial promiscuity (Coleman et al., 2006; Fraser et al., 2007, Vergin et al., 2007). In particular, this study demonstrates the presence of genes within the HVR2 region, which are not present in the genome of ‘Cand. P. ubique’ HTCC1062 or HTCC1002. Therefore, fosmid clone 01–003783 provides further evidence for the genomic plasticity of this bacterial lineage and adaptability to specific environmental conditions, which may have been acquired through horizontal gene transfer.
Accession codes
References
Béjà O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Fraser C, Hanage WP, Spratt BG . (2007). Recombination and the nature of bacterial speciation. Science 315: 476–480.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Rittmann D, Sorger-Herrmann U, Wendisch VF . (2005). Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl Environ Microbiol 71: 4339–4344.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical pacific. PLoS Biol 5: 398–431.
Suzuki MT, Preston CM, Beja O, de la Torre JR, Steward GF, DeLong EF . (2004). Phylogenetic screening of ribosomal RNA gene-containing clones in Bacterial Artificial Chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol 48: 473–488.
Vergin KL, Tripp HJ, Wilhelm LJ, Denver DR, Rappe MS, Giovannoni SJ . (2007). High intraspecific recombination rate in a native population of Candidatus Pelagibacter ubique (SAR11). Environ Microbiol 9: 2430–2440.
Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ . (2007). Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2: 27.
Acknowledgements
This work is supported by a grant from the Natural Environment Research Council (NE/C507902/1) and is part of the core research programme of the Plymouth Marine Laboratory, a collaborative centre of NERC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Gilbert, J., Mühling, M. & Joint, I. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique’ genome. ISME J 2, 790–793 (2008). https://doi.org/10.1038/ismej.2008.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2008.49
Keywords
This article is cited by
-
Comparable light stimulation of organic nutrient uptake by SAR11 and Prochlorococcus in the North Atlantic subtropical gyre
The ISME Journal (2013)
-
High Contribution of SAR11 to Microbial Activity in the North West Mediterranean Sea
Microbial Ecology (2012)
-
Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing
The ISME Journal (2009)
-
Bias in assessments of marine SAR11 biodiversity in environmental fosmid and BAC libraries?
The ISME Journal (2009)