Abstract
Microbial activities drive the global nitrogen cycle, and in the past few years, our understanding of nitrogen cycling processes and the micro-organisms that mediate them has changed dramatically. During this time, the processes of anaerobic ammonium oxidation (anammox), and ammonia oxidation within the domain Archaea, have been recognized as two new links in the global nitrogen cycle. All available evidence indicates that these processes and organisms are critically important in the environment, and particularly in the ocean. Here we review what is currently known about the microbial ecology of anaerobic and archaeal ammonia oxidation, highlight relevant unknowns and discuss the implications of these discoveries for the global nitrogen and carbon cycles.
Similar content being viewed by others
Introduction
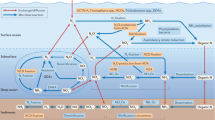
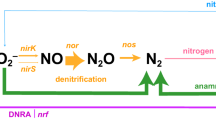
The element nitrogen (N) is an essential nutrient for all organisms, and as a critical component of proteins, N is fundamental to the structures and biochemical processes that define life. N is of such centrality that it has been suggested to be perhaps the best bio-signature for life on other planets (Capone et al., 2006), yet our understanding of how this element is cycled on Earth has changed drastically in just the last few years. Here N exists in multiple oxidation states and chemical forms, and is rapidly converted by micro-organisms on land and in the sea. Until recently, the global N cycle was thought of as essentially ‘linear’ from the atmosphere and back again. The largest reservoir of N on Earth is triple-bonded N2 gas (78% of the atmosphere) and must be fixed by microorganisms before it is readily useable by other organisms. N exists in its most reduced state within organisms, but is rapidly nitrified to nitrate (aerobically) when released following cell death and lysis. Nitrate is in turn denitrified to N2 gas under suboxic to anoxic conditions, completing the cycle (Figure 1).
Microbial nitrogen transformations above, below and across an oxic/anoxic interface in the marine environment (based in part on Arrigo, 2005). Nitrite is highlighted in red to emphasize the central role of this metabolic intermediate/product within and between N-cycling pathways. Key functional genes discussed in the text are shown in yellow: amo, ammonia mono-oxygenase; hao, bacterial hydroxylamine oxidoreductase (?=unknown gene/enzyme in AOA); nir, nitrite reductase; and nor, nitric oxide reductase. For clarity, other functional genes and the process of nitrate/nitrite assimilation are not shown.
Along this flowpath, micro-organisms directly catalyze the processes of nitrification and denitrification, but these two functional groups are a study in contrasts. In the conventional view of nitrification (NH3 → NO2− → NO3−), the metabolic labour is divided between two distinct groups of organisms, ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). Ammonia oxidation is typically thought to be an obligatory aerobic, chemoautotrophic process restricted to just a few groups within the Proteobacteria (Kowalchuk and Stephen, 2001). In contrast, denitrification is primarily heterotrophic, facultative, occurs under low-oxygen conditions and is widespread among over 50 different genera (Zumft, 1997), including members of the Bacteria, Archaea, and – based on the surprising discovery of complete denitrification in a benthic foraminifer (Risgaard-Petersen et al., 2006) – Eukarya. Known denitrifying bacteria and archaea posses several clusters of genes involved in denitrification (Philippot, 2002), and most are therefore capable of performing the multi-step process in its entirety (NO3− → NO2− → NO → N2O → N2).
Much is now known about these processes and many of the micro-organisms involved, yet our understanding of the N cycle has been upended twice in the past few years, first by the discovery of anaerobic ammonium oxidation in natural systems, and more recently by the discovery of aerobic ammonia oxidation within the domain Archaea. Aerobic oxidation of ammonia by bacteria was first discovered over 100 years ago (Winogradsky, 1890), underlining just how rapidly these two major discoveries have taken place. In this review, we focus on recent developments related to the microbial ecology of anaerobic and archaeal ammonia oxidation. We expand upon existing reviews that cover various aspects of the microbial N cycle (Kowalchuk and Stephen, 2001; Zehr and Ward, 2002; Strous and Jetten, 2004; Arrigo, 2005; Kuypers et al., 2006; Nicol and Schleper, 2006; Revsbech et al., 2006), and focus particularly on archaeal ammonia oxidation, because this very recent discovery is an area of remarkably active research.
Anammox
Since the mid-1960s, oceanographers have recognized pervasive ammonium deficits in anoxic basins that hinted at the possible removal of ammonium by anaerobic microbial activity (Richards, 1965). Nevertheless, for the remainder of the century, heterotrophic denitrification was considered the only sink for fixed nitrogen under anoxic conditions in natural systems. The first evidence for anaerobic ammonium oxidation (anammox) to N2 gas was obtained from anoxic (denitrifying) bioreactors of wastewater treatment plants (WWTPs) (Mulder et al., 1995), where it was eventually determined that novel organisms related to Planctomycetales were capable of oxidizing ammonium using nitrite (rather than O2) as the electron acceptor (Strous et al., 1999). Befitting micro-organisms capable of such a novel metabolism, these ‘anammox’ bacteria have a number of truly unique features, including the use of hydrazine (N2H4, i.e., rocket fuel) as a free catabolic intermediate, the biosynthesis of ladderane lipids and the presence of an anammoxosome (intracytoplasmic compartment). All four currently recognized genera of anammox bacteria – Candidatus ‘Brocadia’, ‘Kuenenia’, ‘Scalindula’, and ‘Anammoxoglobus’ – share these unique physiological and morphological features.
Owing to their distinct metabolism and physiology, anammox bacteria received considerable attention in engineered systems, but were assumed to be minor players in the N cycle within natural ecosystems. However, in 2002, Thamdrup and Dalsgaard found anammox to be responsible for 24–67% of N loss in marine sediments (Thamdrup and Dalsgaard, 2002), and in 2003, two parallel studies demonstrated that anammox was directly responsible for a substantial fraction of N loss in the ocean (Dalsgaard et al., 2003; Kuypers et al., 2003). In fact, 20–40% of N loss could be attributed to anammox in the suboxic water columns of the Black Sea and Gulfo Dulce (Dalsgaard et al., 2003; Kuypers et al., 2003), and more recent studies indicate that, if anything, anammox is responsible for an even greater percentage of marine N loss (Kuypers et al., 2005; Hamersley et al., 2007). The now well-established significance of anammox in the marine environment has emerged from a combination of 15N-based tracer studies, analysis of ladderane lipid biomarkers, fluorescent in situ hybridization and phylogenetic and quantitative PCR analysis of 16S rRNA sequences (Schmid et al., 2005). To date, anammox has been documented in marine, coastal and estuarine sediments (Thamdrup and Dalsgaard, 2002; Trimmer et al., 2003; Risgaard-Petersen et al., 2004; Rysgaard et al., 2004; Engstrom et al., 2005), anoxic basins (Dalsgaard et al., 2003; Kuypers et al., 2003), oxygen minimum zones (OMZs) off of West Africa, Chile and Peru (Kuypers et al., 2005; Thamdrup et al., 2006; Hamersley et al., 2007), mangroves (Meyer et al., 2005), sea-ice (Rysgaard and Glud, 2004) and freshwater lakes – including Lake Tanganyika, the second largest lake in the world (Schubert et al., 2006). A recent PCR-based study using newly designed primers detected ‘Scalindula’-like 16S rRNA genes in a variety of freshwater and marine sediments, as well as permafrost soil (Penton et al., 2006). Although anammox activity has yet to be measured in soils, the occurrence of anammox in this broad array of aquatic environments suggests that this process is truly ubiquitous, and it seems likely that anammox will be found in virtually any N-containing ecosystem with a pronounced suboxic zone or chemocline.
In addition to their widespread distribution, anammox bacteria appear to be far more metabolically versatile than previously thought: these organisms have the capacity to couple the oxidation of various organic acids to the reduction of nitrate (Guven et al., 2005); Candidatus ‘Anammoxoglobus propionicus’ outcompeted other anammox bacteria and heterotrophic denitrifiers for the oxidation of propionate in the presence of ammonium, nitrite and nitrate (Kartal et al., 2007b); the capacity to respire iron and manganese oxides was suggested by the large number of encoded c-type cytochromes in the Kuenenia stuttgartiensis genome (by analogy to the metal-respiring bacteria Geobacter and Shewanella), and this ability was confirmed experimentally (Strous et al., 2006). In addition, K. stuttgartiensis is capable of dissimilatory nitrate reduction to ammonium (DNRA), reducing NO3– to NH4+ even in the presence of 10 mM NH4+ (Kartal et al., 2007a). Regardless of whether DNRA is performed by anammox or other (facultatively) anaerobic micro-organisms in situ, DNRA could provide NH4+ for anammox, and because this NH4+ is eventually lost as N2 gas, the whole process is effectively concealed as denitrification – in other words, an even larger fraction of N loss from anoxic systems may be driven by anammox organisms. In fact, evidence for DNRA has been detected in the Benguela upwelling system (Kartal et al., 2007a), where anammox bacteria actively remove massive amounts of N (Kuypers et al., 2005).
Coupled DNRA–anammox is indistinguishable from denitrification based on most conventional isotope tracer experiments, such that more sophisticated tracer methods and/or gene marker studies are needed to assess the importance of this process in the environment. Although a calcium-dependent cytochrome c protein with a high rate of nitrite reduction to ammonium was partially purified from K. stuttgartiensis, and candidate genes for this enzyme were putatively identified in the K. stuttgartiensis genome (Kartal et al., 2007a), there is currently no established functional gene marker for analyzing anammox bacteria in the environment. This is in clear contrast to other N-cycling micro-organisms. For example, functional genes encoding nitrite reductase (nirK and nirS) and ammonia mono-oxygenase (amoA) are commonly used molecular markers for studying communities of denitrifiers and aerobic ammonia oxidizers, whereas only 16S rRNA genes have been used to detect anammox bacteria. However, functional genes encoding the most defining metabolic feature of anammox – hydrazine metabolism – are beginning to be identified: candidate hydrazine hydrolase and hydrazine dehydrogenase genes have been identified in the K. stuttgartiensis genome (Strous et al., 2006), and a hydrazine-oxidizing enzyme (HZO) and the underlying genes have recently been identified in a related organism (Shimamura et al., 2007). Once these functional genes are definitively and specifically linked to anammox, analysis of anammox functional gene abundance and expression in the environment becomes a real possibility.
At the global scale, the overall contribution of anammox to global N loss is still unclear. On land, few if any measurements of anammox activity exist, but the widespread occurrence of anammox in marine sediments and suboxic water columns indicates that this process could be responsible for a substantial proportion of fixed N loss from the ocean. In marine sediments, the proportion of N loss that can be attributed to anammox appears to increase with water depth (Thamdrup and Dalsgaard, 2002; Trimmer et al., 2003; Risgaard-Petersen et al., 2004; Rysgaard et al., 2004; Engstrom et al., 2005); this suggests anammox may be important in slope and hemipelagic sediments, where modeling results indicate that the majority of benthic N losses take place (Middelburg et al., 1996). A substantial portion of marine N loss occurs in oceanic water columns, specifically in spatially-constrained OMZs over a relatively limited depth range. To the best of our knowledge, there is so far little published evidence from 15N-labelling experiments that nitrate is directly converted into N2 by heterotrophic denitrifiers in these regions, whereas anammox appears to dominate N2 production in all published experiments to date (Kuypers et al., 2005; Thamdrup et al., 2006; Hamersley et al., 2007). Altogether, these studies indicate that anammox is probably responsible for 30–50% of all marine N loss (Kuypers et al., 2006).
Ammonia-oxidizing archaea
As anammox continues to be explored as a ‘new’ process in the N cycle, new ‘players’ in the N cycle have also been identified, and they are among the most widely distributed and abundant groups of micro-organisms on the planet – the mesophilic Crenarchaeota. Although archaea were previously characterized as extremophiles, mesophilic archaea are now recognized to be an ubiquitous component of marine plankton (DeLong, 1992; Fuhrman et al., 1992), with the marine ‘group 1’ clade of Crenarchaeota alone comprising over 20% of picoplankton in the world ocean (Karner et al., 2001). These organisms are estimated to number a staggering 1028 cells in total; however, because of our inability to cultivate them, for the last 15 years our understanding of their physiology and biogeochemical function remained almost entirely speculative. Remarkably, two complementary metagenomic studies of seawater (Venter et al., 2004) and soil (Treusch et al., 2005) revealed putative ammonia mono-oxygenase (amoA) genes derived from uncultivated Crenarchaeota, suggesting the genetic capacity for ammonia oxidation. More specifically, Venter et al. identified an amoA-like gene on an archaeal-associated scaffold, whereas Treusch et al. found a similar gene on the same 43-kb soil DNA fragment as a 16S rRNA gene derived from a member of the group 1.1b Crenarchaeota – the most widespread crenarchaeal group in soils (Ochsenreiter et al., 2003).
The definitive link between these novel amoA genes and archaeal ammonia oxidation was recently and convincingly established by cultivation of an ammonia-oxidizing crenarchaeon – designated Nitrosopumilus maritimus – from a saltwater aquarium (Könneke et al., 2005). N. maritimus grows chemoautotrophically to cell densities of 107 ml−1 via the near-stoichiometric conversion of ammonia into nitrite, and with bicarbonate as a sole carbon source (organic carbon actually inhibited growth) (Könneke et al., 2005). 16S rRNA phylogeny places this organism firmly in the group 1.1a Crenarchaeota – the first cultivated representative from this exceptionally abundant archaeal group – and it contains putative ammonia mono-oxygenase genes amoA, amoB and amoC.
Hot on the heels of this discovery came the first molecular evidence demonstrating the archaeal amoA gene to be pervasive in areas of the ocean that are critical for the global nitrogen cycle – including the base of the euphotic zone, suboxic water columns and coastal/estuarine sediments (Francis et al., 2005). For the first time, these data indicated that many marine Crenarchaeota might be capable of ammonia oxidation, and these organisms were identified as putative ammonia-oxidizing archaea (AOA) (Francis et al., 2005). In addition, phylogenetic analysis of several hundred archaeal amoA sequences revealed diverse and distinct AOA communities associated with different habitats and sampling sites, with little overlap between water columns and sediments. Considering that only a few genera of Bacteria (Nitrosomonas, Nitrosospira and Nitrosococcus) were thought to be involved in ammonia oxidation (Purkhold et al., 2000), this diversity among AOA is all the more remarkable.
Following these initial insights, our picture of AOA in the environment has become more complete and more compelling. For example, AOA appear to be active in natural samples, based on quantification of amoA gene expression in soil microcosms, specifically increased amoA expression in the presence of elevated ammonia concentrations (Treusch et al., 2005). Metagenomic analysis of the only other established species within the marine group 1.1a Crenarchaeota, the uncultivated sponge symbiont Cenarchaeum symbiosum, identified putative ammonia mono-oxygenase genes (amoA, amoB, amoC), as well as homologs of ammonia permease, urease, a urea transport system, putative nitrite reductase and nitric oxide reductase accessory protein – all potentially associated with chemoautotrophic ammonia oxidation (Hallam et al., 2006a, 2006b). Surprisingly, homologs for critical components of the second enzymatic step of bacterial ammonia oxidation – hydroxylamine oxidoreductase and cytochromes c554 and c552 – were not identified (Hallam et al., 2006b), indicating that if C. symbiosum is in fact capable of ammonia oxidation, it apparently employs a different mechanism than known AOB for catalyzing this key reaction (Hallam et al., 2006a). This has been interpreted by some as evidence that archaeal ammonia oxidation evolved fairly late by incorporating an AMO-like biochemical function into an ammonia-independent metabolism (Klotz et al., 2006).
Multiple components of a modified 3-hydroxypropionate cycle (for autotrophic carbon assimilation) – as well as genes predicted to encode a nearly complete oxidative tricarboxylic acid cycle (consistent with organic carbon consumption) – were also present in C. symbiosum, suggesting that this organism has the potential to function either as a strict autotroph, or as a mixotroph utilizing both CO2 and organic material as carbon sources (Hallam et al., 2006a, 2006b). In fact, organic geochemical evidence indicate that either or both of these metabolic lifestyles are real possibilities. Previous studies have demonstrated uptake of organic compounds by archaea in the form of amino acids (Ouverney and Fuhrman, 2000; Herndl et al., 2005; Teira et al., 2006), whereas signatures of chemoautotrophy have been identified among the archaea based on the carbon isotopic composition of archaeal membrane lipids and remains (Kuypers et al., 2001; Pearson et al., 2001), and the direct incorporation of 13C-labeled bicarbonate into crenarchaeal lipids in the absence of light (Wuchter et al., 2003). Most recently, Ingalls et al. used the natural distribution of radiocarbon in archaeal membrane lipids to quantify the bulk carbon metabolism of archaeal communities at two depths in the subtropical North Pacific gyre. In surface waters, archaea incorporated modern carbon into their membrane lipids, whereas archaea at 670 m incorporated carbon that was isotopically enriched relative to inorganic carbon at the same depth (Ingalls et al., 2006). On the basis of an isotopic mass balance model, 83% of in situ archaeal production at 670 m was estimated to be chemoautotrophic, and the balance heterotrophic. It remains to be determined whether this is indicative of archaeal mixotrophy, or a mixed community of autotrophic and heterotrophic archaea dominated by autotrophs.
Given the sheer numbers of Crenarchaeota in the ocean and evidence that many are AOA, it is not surprising that AOA appear to be much more abundant than AOB (Leininger et al., 2006; Wuchter et al., 2006; Mincer et al., in press), which typically comprise <0.1% of microbial communities (Ward, 2000). In the North Sea and North Atlantic, archaeal amoA gene copy numbers were 10–1000 times those of betaproteobacterial amoA and correlated with cell counts of Crenarchaeota (Wuchter et al., 2006). In a study of 12 pristine and agricultural soils spanning three climate zones, archaeal amoA gene copy numbers were up to 3000 times those of the betaproteobacteria and correlated with Crenarchaeota-specific lipids, including crenarchaeol (Leininger et al., 2006). Reverse transcription quantitative PCR studies and complementary DNA analysis demonstrated the expressional activity of AOA in situ, and supported the numerical dominance of AOA over AOB. Together, these findings suggest that most mesophilic Crenarchaeota are AOA, and that these organisms are the numerically dominant ammonia oxidizers in the ocean and in soils.
Adding to this emerging paradigm of AOA ubiquity, AOA have also recently been detected in nitrifying wastewater treatment bioreactors (Park et al., 2006), where 50 of 75 archaeal amoA sequences recovered in activated sludge from Oregon, Wisconsin, Pennsylvania and New Jersey were virtually identical. In estuarine sediments from Bahía del Tóbari, Mexico, AOA communities from the interior of the estuary were phylogenetically distinct from those found at the mouths of the estuary, and the distribution of these two archaeal amoA ‘ecotypes’ was consistent with amoA genes being widespread within both group 1.1a and 1.1b Crenarchaeota (Beman and Francis, 2006). Closely related archaeal amoA sequences were recently recovered from an Austrian radioactive thermal spring (Weidler et al., 2007) and a Colorado geothermal mine adit (Spear, Barton, Robertson, Francis and Pace, unpublished results), which suggests AOA may be important in subsurface/cave ecosystems, and could be indicative of crenarchaeal ammonia oxidation at higher temperatures (for example, 45–50°C). Although these subsurface environments are not truly ‘hyperthermophilic’, non-marine crenarchaeol has been detected in Nevada hot springs with temperatures from 40°C to 84°C (Pearson et al., 2004; Zhang et al., 2006), and archaeal amoA genes also seem to occur widely in terrestrial hot springs up to 86°C (Zhang et al., unpublished results). Reports of N-fixation in 53.5-63.4°C hot spring cyanobacterial mats (Steunou et al., 2006), and at 92°C in a deep-sea hydrothermal vent methanogen (Mehta and Baross, 2006), may be indicative of active N cycling in high-temperature environments. At this stage, it is unclear whether amoA genes are also associated with some thermophilic Crenarchaeaota lineages, or whether the thermotolerance and ecological niche of ‘mesophilic’ Crenarchaeota is more extensive than currently thought; both are certain to be areas of active research.
Recent results from the North Pacific Ocean add at least one piece to this puzzle. Here Mincer et al., (in press) found discrepancies between archaeal amoA gene copy numbers and crenarchaeal 16S rRNA gene copy numbers, where amoA was several orders of magnitude more abundant than 16S rRNA at certain depths. However, when 16S rRNA genes corresponding to the deeply branching pSL12 clade (originally discovered in a Yellowstone hot spring; Barns et al., 1996) were specifically quantified, crenarchaeal amoA and 16S rRNA gene copy numbers were more comparable. This intriguing finding suggests that the pSL12 clade – which has only been reported a few times previously in the marine environment (Vetriani et al., 1999; van der Wielen et al., 2005; Zaballos et al., 2006) – may be widespread and at times abundant in the ocean. These data can also be interpreted as evidence for amoA within the pSL12 clade, potentially adding to the list of crenarchaeal groups known to be capable of ammonia oxidation (Mincer et al., in press). Finally, corroborating a growing number of studies, direct comparison of AOA and AOB abundance in the North Pacific Gyre and Monterey Bay showed AOA to be up to two orders of magnitude more abundant than their often undetectable betaproteobacterial counterparts (Mincer et al., in press).
New paradigms
Our understanding of these new processes and players in the microbial N cycle has evolved in opposite directions for anammox and the AOA. In the case of anammox, initial observations based on biogeochemistry led to the discovery of these organisms in the environment, and we are only now beginning to determine the biochemical pathways and genes involved in anammox. AOA were first identified via functional gene sequences recovered directly from the environment, and we still do not know what their full contributions are to N biogeochemistry.
The lesson to be learned from this is that neither of these processes can be fully characterized without employing multiple, complimentary approaches – including molecular approaches, metagenomics, cultivation and (bio)geochemistry – to address a growing number of questions. For example, how do the diversity, abundance and activity of these organisms compare with their presumed competitors, the denitrifiers and AOB? Anammox is the dominant N loss process in several OMZs, but is this true for all? Are there times and places in the ocean where conventional denitrification dominates N loss? And why does denitrification appear to be of greater significance in sediments and lakes? Similarly, AOA are far more abundant than AOB in a number of environments – are there exceptions? Is this true with regard to their activity? Are these organisms truly ‘functionally equivalent’? Do AOA, AOB and anammox bacteria and their respective ‘ecotypes’ respond differently to environmental perturbations and gradients (for example N, oxygen, light, salinity)?
Ultimately, a new N cycle paradigm will only emerge through considering all of these processes and microbial groups together, particularly in their use of common substrates and in environments where they coexist. For example, AOA, AOB, anammox bacteria and denitrifiers all appear to possess nitrite reductase (nirK/S) genes (Casciotti and Ward, 2001; Treusch et al., 2005; Strous et al., 2006; Hallam et al., 2006b; Cantera and Stein, 2007). Although in some cases these genes may only be involved in nitrite detoxification, this parallel suggests that nitrite could act as an important substrate for all of these organisms. More broadly, if most of the 1028 Crenarchaeota cells in the ocean are capable of ammonia oxidation (as all available evidence indicates), and do so even at fairly low rates, the fact that nitrite only rarely accumulates in the ocean necessitates a large and active pool of nitrite-oxidizing organisms. In direct support of this idea, correlation between the quantitative depth distributions of planktonic crenarchaea and 16S rRNA sequences similar to those of known nitrite-oxidizing Nitrospina species suggests metabolic coupling between AOA and NOB (via nitrite) in the ocean (Mincer et al., in press).
The distribution of the radiatively active trace gas nitrous oxide (N2O) may also be strongly influenced by these organisms: AOB (Casciotti and Ward, 2005), denitrifiers, and now AOA (Hallam et al., 2006b) possess nitric oxide reductase (nor) genes that could be involved in N2O production. Anammox bacteria are now known to produce N2O as well (Kartal et al., 2007a) – not as an intermediate of the anammox reaction but apparently as a result of NO detoxification. Even without AOA and anammox in the mix, the sources of N2O are highly uncertain; with two new potential contributors, our understanding of how, where and why N2O is produced may need rethinking.
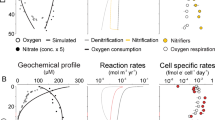
Production of N2O is likely to be of particular importance under suboxic conditions, and here our understanding of anammox and the AOA – and the N cycle in general – is at its most muddled. The fact that anammox appears to dominate N loss in OMZs seriously challenges our understanding of organic matter remineralization in these regions. Without remineralization of NH4+ from organic matter via denitrification, it is unclear how anammox could be sustained; one possibility is that microaerobic heterotrophs are responsible for regenerating N under low, but non-zero, oxygen conditions commonly found in OMZs. Although AOA are presumably aerobic ammonia oxidizers, AOA (Francis et al., 2005; Coolen et al., 2007) and the archaea generally (Kuypers et al., 2001; Sinninghe Damste et al., 2002), appear to be remarkably successful under low oxygen conditions. We know anammox bacteria and AOA are found at the same depths in the Black Sea (Kuypers et al., 2003; Francis et al., 2005; Kirkpatrick et al., 2006; Coolen et al., 2007), and we assume elsewhere; do they compete for dissolved NH4+ under low oxygen, or are there potentially beneficial interactions among these organisms via the supply of critical substrates, such as nitrite? For example, in the CANON (Completely Autotrophic Nitrogen removal Over Nitrite) process (Third et al., 2001), AOB provide nitrite and also create anoxic microenvironments for anammox bacteria (via O2 consumption) – do AOA play a similar co-operative role in natural systems? Correlations between AOA and anammox bacteria in OMZs – similar to those observed between Archaea and NOB (Mincer et al., in press) – would be good evidence of this and should be explored. For example, Coolen et al. (2007) found archaeal amoA gene copies to be highest at the same suboxic depth where anammox bacteria were also abundant in the Black Sea (Kuypers et al., 2003) (see Figure 2). Surprisingly, crenarchaeol concentrations were highest in the sulfidic zone of the Black Sea. These lipids may be derived from crenarchaeal remains sinking through the water column, however archaeal communities recovered in this zone were very different from those found at other depths, suggestive of active growth (Coolen et al., 2007). There are a number of possible interpretations of these data, but it does seem clear that marine Crenarchaeota have considerable metabolic and ecological flexibility, based on their presence under sulfidic conditions and evident success in suboxic environments.
Depth profiles from the Black Sea of (a) oxygen (black line) and sulphide (white line); (b) the anammox ladderane lipid FAME 1; (c) Crenarchaeota (marine group 1.1a Crenarchaeota 16S rRNA genes as a percent of total archaeal 16S rRNA genes); (d) AOA (archaeal amoA genes as a percent of total archaeal 16S rRNA genes); and (e) the (cren)archaeal membrane lipid crenarchaeol. The suboxic zone (O2<3 μ M) is indicated by light grey shading, and the sulfidic zone (H2S>0 μ M) by dark gray shading. Dashed line indicates depth of nitrite maximum (NO2−>0.3 μ M). Similarities and differences in the distributions of anaerobic and archaeal ammonia oxidizers may or may not be indicative of direct metabolic coupling. Data replotted from Kuypers et al. (2003) and Coolen et al. (2007).
Here, too, the interplay between N cycle and the carbon cycle becomes particularly salient. If N loss in OMZs is principally driven by the autotrophic process of anammox rather than heterotrophic denitrification, what happens to organic carbon, if it is not remineralized via denitrification? With regard to the archaea, their exceptional numbers mean that even if they perform chemoautrophic ammonia oxidation at relatively low rates, they still represent a substantial source of carbon in the deep ocean (Herndl et al., 2005; Ingalls et al., 2006) – is this truly the case? Although answers to some of these questions have already begun to emerge, it is clear that the microbial ecology of anammox bacteria and AOA will be an area of active research for years to come, and will be essential to our understanding of the global N and carbon cycles going forward.
References
Arrigo K . (2005). Marine micro-organisms and global nutrient cycles. Nature 437: 349–355.
Barns SM, Delwiche CF, Palmer JD, Pace NR . (1996). Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA 93: 9188–9193.
Beman JM, Francis CA . (2006). Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hyper-nutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl Environ Microbiol 72: 7677–7777.
Cantera JJL, Stein JL . (2007). Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ Microbiol 9: 765–776.
Capone DG, Popa R, Flood B, Nealson KH . (2006). Follow the Nitrogen. Science 312: 708–709.
Casciotti KL, Ward BB . (2001). Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl Environ Microbiol 67: 2213.
Casciotti KL, Ward BB . (2005). Phylogenetic analysis of nitric oxide reductase gene homologues from aerobic ammonia-oxidizing bacteria. FEMS Microbiol Ecol 52: 197–205.
Coolen MJL, Abbas B, Van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG et al. (2007). Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol 9: 1001–1016.
Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuna-Gonzalez J . (2003). N2 production by the anammox reaction in the water column of Golfo Dulce, Costa Rica. Nature 422: 606–608.
DeLong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
Engstrom P, Dalsgaard T, Hulth S, Aller RC . (2005). Anaerobic ammonium oxidation by nitrite (anammox): implications for N2 production in coastal marine sediments. Geochim Cosmochim Acta 69: 2057–2065.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Fuhrman JA, McCallum K, Davis AA . (1992). Novel major archaebacterial group from marine plankton. Nature 356: 148–149.
Guven D, Dapena A, Kartal B, Schmid MC, Maas B, van de Pas-Schoonen K et al. (2005). Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 71: 1066–1071.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-i, Sugahara J et al. (2006a). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. PNAS 103: 18296–18301.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006b). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol 4: e95.
Hamersley MR, Lavik G, Woebken D, Rattray JE, Lam P, Hopmans EC et al (2007). Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol Oceanog 52 (in press).
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A et al. (2005). Contribution of archaea to total prokaryotic production in the deep atlantic ocean. Appl Environ Microbiol 71: 2303–2309.
Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM et al. (2006). Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA 103: 6442–6447.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Kartal B, Kuypers MMM, Lavik G, Schalk J, Op den Camp HJM, Jetten MSM et al. (2007a). Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ Microbiol 9: 635–642.
Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI et al. (2007b). Candidatus ‘Anammoxoglobus propionicus’ a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Systematic Appl Microbiol 30: 39–49.
Kirkpatrick J, Oakley B, Fuchsman C, Srinivasan S, Staley JT, Murray JW . (2006). Diversity and distribution of planctomycetes and related bacteria in the suboxic zone of the black sea. Appl Environ Microbiol 72: 3079–3083.
Klotz MG, Arp DJ, Chain PSG, El-Sheikh AF, Hauser LJ, Hommes NG et al. (2006). Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium nitrosococcus oceani ATCC 19707. Appl Environ Microbiol 72: 6299–6315.
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Kowalchuk GA, Stephen JR . (2001). Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55: 485–529.
Kuypers MMM, Blokker P, Erbacher J, Kinkel H, Pancost RD, Schouten S et al. (2001). Massive expansion of marine archaea during a mid-Cretaceous anoxic event. Science 293: 92–94.
Kuypers MMM, Lavik G, Thamdrup B . (2006). Anaerobic ammonium oxidation in the marine environment. In: Neretin, LN (ed). Past and Present Water Column Anoxia. Springer: Dordrecht, The Netherlands, pp 311–335.
Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R et al. (2005). Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci 102: 6478–6483.
Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jørgensen BB, Kuenen JG et al. (2003). Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422: 608–611.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806.
Mehta MP, Baross JA . (2006). Nitrogen Fixation at 92°C by a Hydrothermal Vent Archaeon. Science 314: 1783–1786.
Meyer RL, Risgaard-Petersen N, Allen DE . (2005). Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl Environ Microbiol 71: 6142–6149.
Middelburg JJ, Soetaert K, Herman PMJ, Heip CHR . (1996). Denitrification in marine sediments: a model study. Global Biogeochem Cy 10: 661–673.
Mincer TJ, Church MJ, Taylor LT, Preston CM, Karl DM, DeLong EF . (in press). Quantitative distribution of presumptive archaeal bacterial nitrifiers in Monterey Bay the North Pacific Subtropical Gyre. Environ Microbiol. Advance online publication, 12 February 2007; doi:10.1111/j.1462-2920.2006.01227.x.
Mulder A, Graaf AA, Robertson LA, Kuenen JG . (1995). Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16: 177–184.
Nicol GW, Schleper C . (2006). Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14: 207–212.
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C . (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5: 787.
Ouverney CC, Fuhrman JA . (2000). Marine planktonic archaea take up amino acids. Applied Environ Microbiol 66: 4829–4833.
Park H-D, Wells GF, Bae H, Criddle CS, Francis CA . (2006). Occurrence of ammonia-oxidizing Archaea in wastewater treatment plant bioreactors Appl. Environ Microbiol 72: 5643–5647.
Pearson A, Huang Z, Ingalls AE, Romanek CS, Wiegel J, Freeman KH et al. (2004). Nonmarine crenarchaeol in Nevada hot springs. Appl Environ Microbiol 70: 5229–5237.
Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI . (2001). Origns of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound-specific 14C analysis. Geochimi Cosmochim Acta 65: 3123–3137.
Penton CR, Devol AH, Tiedje JM . (2006). Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72: 6829–6832.
Philippot L . (2002). Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta 1577: 355–376.
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops H-P, Wagner M . (2000). Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66: 5368–5382.
Revsbech NP, Risgaard-Petersen N, Schramm A, Nielsen LP . (2006). Nitrogen transformations in stratified aquatic microbial ecosystems. Antonie van Leeuwenhoek 90: 361–375.
Richards FA . (1965). Anoxic basins and fjords. In: Ripley, JP and Skirrow G (eds). Chemical Oceanography. Academic Press: London, pp 611–645.
Risgaard-Petersen N, Langezaal AM, Ingvardsen S, Schmid MC, Jetten MSM, Op den Camp HJM et al. (2006). Evidence for complete denitrification in a benthic foraminifer. Nature 443: 93–96.
Risgaard-Petersen N, Meyer RL, Schmid M, Jetten MSM, Enrich-Prast A, Rysgaard S et al. (2004). Anaerobic ammonium oxidation in an estuarine sediment. Aquatic Microbial Ecol 36: 293–304.
Rysgaard S, Glud RN . (2004). Anaerobic N2 production in Arctic sea ice. Limnol Oceanog 49: 86–94.
Rysgaard S, Glud RN, Risgaard-Petersen N, Dalsgaard T . (2004). Denitrification and anammox activity in Arctic marine sediments. Limnol Oceanog 49: 1493–1502.
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B et al. (2005). Biomarkers for in situ detection of anaerobic ammonium-oxidizing (Anammox) bacteria. Appl Environ Microbiol 71: 1677–1684.
Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MMM . (2006). Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ Microbiol 8: 1857–1863.
Shimamura M, Nishiyama T, Shigetomo H, Toyomoto T, Kawahara Y, Furukawa K et al. (2007). Isolation of a multiheme protein from an anaerobic ammonium-oxidizing enrichment culture with features of a hydrazine-oxidizing enzyme. Appl Environ Microbiol 73: 1065–1072.
Sinninghe Damste JS, Rijpstra WIC, Hopmans EC, Prahl FG, Wakeham SG, Schouten S . (2002). Distribution of membrane lipids of planktonic crenarchaeota in the arabian sea. Appl Environ Microbiol 68: 2997.
Steunou A-S, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E et al. (2006). In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA 103: 2398–2403.
Strous M, Jetten MSM . (2004). Anaerobic oxidation of methane and ammonium. Annu Rev Microbiol 58: 99–117.
Strous M, Kuenen JG, Jetten MSM . (1999). Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol 65: 3248–3250.
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440: 790–794.
Teira E, van Aken H, Veth C, Herndl GJ . (2006). Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol Oceanography 51: 60–69.
Thamdrup B, Dalsgaard T . (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol 68: 1312–1318.
Thamdrup B, Dalsgaard T, Jensen MM, Ulloa O, Farias L, Escribano R . (2006). Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol Oceanog 51: 2145–2156.
Third KA, Sliekers AO, Kuenen JG, Jetten MSM . (2001). The CANON system (completely autotrophic nitrogen removal over nitrite) under ammonium limitation: interaction and competition between three groups of bacteria. Syst Appl Microbiol 24: 588–596.
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk H-P, Schleper C . (2005). Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7: 1985–1995.
Trimmer M, Nicholls JC, Deflandre B . (2003). Anaerobic ammonium oxidation measured in sediments along the Thames estuary, United Kingdom. Appl Environ Microbiol 69: 6447–6454.
van der Wielen PWJJ, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L et al. (2005). The Enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307: 121–123.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Vetriani C, Jannasch H, MacGregor B, Stahl D, Reysenbach A . (1999). Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol 65: 4375–4384.
Ward BB . (2000). Nitrification and the marine nitrogen cycle. In: Kirchman D (ed). Microbial Ecology of the Oceans. Wiley and Sons: New York, pp 427–454.
Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H . (2007). Communities of archaea and bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl Environ Microbiol 73: 259–270.
Winogradsky S . (1890). 2nd memoire. Recherches sur les organismes de la nitrification. Ann Inst Pasteur 4: 257–275.
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P et al. (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322.
Wuchter C, Schouten S, Boschker HTS, Sinnighe Damsté JS . (2003). Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett 219: 203–207.
Zaballos M, Lopez-Lopez A, Ovreas L, Bartual SG, D’Auria G, Alba JC et al. (2006). Comparison of prokaryotic diversity at offshore oceanic locations reveals a different microbiota in the Mediterranean Sea. FEMS Microbiol Ecol 56: 389–405.
Zehr JP, Ward BB . (2002). Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl Environ Microbiol 68: 1015–1024.
Zhang CL, Pearson A, Li YL, Mills G, Wiegel J . (2006). Thermophilic temperature optimum for crenarchaeol synthesis and its implication for archaeal evolution. Appl Environ Microbiol 72: 4419–4422.
Zumft WG . (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616.
Acknowledgements
We thank Tracy Mincer and Ed DeLong for generously providing their manuscript prior to publication, John Spear and Chuanlun Zhang for sharing unpublished results, and Alyson Santoro and several anonymous reviewers for useful comments on the manuscript. MMMK thanks Gaute Lavik, Bo Barker Jørgensen, Bo Thamdrup, Bess Ward, and Marc Strous for discussions. CAF was supported in part by National Science Foundation MIP grant MCB-0604270, JMB was supported in part by National Science Foundation grant OCE-0623575 (to Jed Fuhrman), and MMMK was funded by the Max Planck Society.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Francis, C., Beman, J. & Kuypers, M. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1, 19–27 (2007). https://doi.org/10.1038/ismej.2007.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.8
Keywords
This article is cited by
-
N/S element transformation modulating lithospheric microbial communities by single-species manipulation
Microbiome (2023)
-
Nitrification and beyond: metabolic versatility of ammonia oxidising archaea
The ISME Journal (2023)
-
Contribution of ammonia-oxidizing archaea and bacteria to nitrogen transformation in a soil fertilized with urea and organic amendments
Scientific Reports (2023)
-
Microbial communities in the deep-sea sediments of the South São Paulo Plateau, Southwestern Atlantic Ocean
International Microbiology (2023)
-
Vertical microbial profiling of water column reveals prokaryotic communities and distribution features of Antarctic Peninsula
Acta Oceanologica Sinica (2023)