Abstract
The use of antibiotics at subtherapeutic concentrations for agricultural applications is believed to be an important factor in the proliferation of antibiotic-resistant bacteria. The goal of this study was to determine if the application of manure onto agricultural land would result in the proliferation of antibiotic resistance among soil bacteria. Chlortetracycline-resistant bacteria were enumerated and characterized from soils exposed to the manure of animals fed subtherapeutic concentrations of antibiotics and compared to the chlortetracycline-resistant bacteria from soils at farms with restricted antibiotic use (dairy farms) and from non-agricultural soils. No significant differences were observed at nine different study sites with respect to the numbers and types of cultivated chlortetracycline-resistant bacteria. Genes encoding for tetracycline resistance were rarely detected in the resistant bacteria from these sites. In contrast, soils collected from a tenth farm, which allowed manure to indiscriminately accumulate outside the animal pen, had significantly higher chlortetracycline-resistance levels. These resistant bacteria frequently harbored one of 14 different genes encoding for tetracycline resistance, many of which (especially tet(A) and tet(L)) were detected in numerous different bacterial species. Subsequent bacterial enumerations at this site, following the cessation of farming activity, suggested that this farm remained a hotspot for antibiotic resistance. In conclusion, we speculate that excessive application of animal manure leads to the spread of resistance to soil bacteria (potentially by lateral gene transfer), which then serve as persistent reservoir of antibiotic resistance.
Similar content being viewed by others
Introduction
Antibiotics play an important role in controlling infectious disease. Many antibiotics used in clinical practice are naturally occurring, such that resistance to antibiotics among bacteria predates their modern clinical use. The past few decades, however, have witnessed a steady increase in the number and diversity of antibiotic-resistant bacteria – rendering some bacterial infections virtually untreatable (Livermore, 2003). The cost of resistance associated with the treatment of infections caused by antibiotic-resistant bacteria has increased many-fold (Levy and Marshall, 2004). The pervasive use of antibiotics for both therapeutic and non-therapeutic purposes has been blamed for the widespread occurrence of antibiotic-resistant bacteria, because the use of antibiotics creates a selective pressure in favor of resistant bacteria.
Agricultural use accounts for at least half of the antibiotics produced in the United States (Lipsitch et al., 2002). Antibiotics are used in agriculture for the treatment of sick animals, disease prophylaxis, growth promotion and crop dusting. Growth promotion involves the addition of subtherapeutic doses of antibiotics to animal feed to induce faster weight gain, whereas prophylaxis involves antibiotic use to prevent disease outbreaks. There is considerable controversy regarding the necessity of such subtherapeutic antibiotic use compared to the threat it poses to human health (Ferber, 2003; Livermore, 2003; Arnold et al., 2004; Phillips et al., 2004). While it is agreed that subtherapeutic antibiotic use leads to an increase in antibiotic-resistant fecal bacteria in the animals (Levy, 1978; Aarestrup et al., 2001; Hayes et al., 2004; Jackson et al., 2004), the role of subtherapeutic antibiotic use in the global spread of antibiotic resistance remains ambiguous.
The disposal of animal waste could be a possible route for the spread of antibiotic resistance from animal farms. Animal waste generated at farms is typically applied to agricultural fields as a fertilizer; however, many animal farms are too large compared to the quantity of available land (United States Environment Protection Agency, 2004). This problem has become more pertinent over the last few decades because of a gradual transition from small farms to concentrated animal feeding operations (CAFOs). Thus, there is an increased risk of adding persistent antibiotic residues and resistant fecal bacteria to soil, potentially leading to the proliferation of resistance among indigenous bacteria (Chee-Sanford et al., 2001; De Liguoro et al., 2003; Sørum et al., 2006).
While numerous studies have focused on the antibiotic resistance and survival of specific commensal bacteria (for example, Enterococci) originating from farms using subtherapeutic concentrations of antibiotics in their animal feed (for examples, see Aarestrup et al., 2001, 2002 ; Hayes et al., 2004), few studies have investigated the proliferation of antibiotic resistance among diverse species of bacteria (for examples, see Aminov et al., 2002; Sengelov et al., 2003). In the present study, antibiotic-resistant bacteria were enumerated and characterized from soils fertilized with manure at farms using subtherapeutic concentrations of antibiotics in their animal feed. We hypothesized that the application of manure from farms using subtherapeutic antibiotics on agricultural land would result in the proliferation of antibiotic resistance among the indigenous soil bacteria. As a basis of comparison, antibiotic-resistant bacteria were also enumerated and characterized from soils fertilized with manure from dairy farms (DFs) (where antibiotic use is more restricted) and from non-agricultural soils. Our results suggest that excessive application of animal manure leads to the spread of resistance to soil bacteria (potentially by lateral gene transfer), which then serve as persistent reservoir of antibiotic resistance.
Methods
Study sites and sample collection
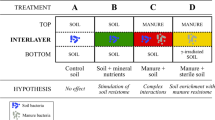
Soil samples from 10 sites were studied, including four swine feeding operations (SFOs), three dairy farms and three non-agricultural areas (NAGs) (Table 1). Site SFO1 was a small swine farm, where animal waste was allowed to overflow the animal pen and accumulate. The other three SFOs were large CAFOs (>1000 animal units per year) housing pigs, where manure was stored in underground pits for 6–12 months before land application. At these sites, liquid manure was injected into agricultural fields approximately six inches below the soil surface, once every 2–4 years. Antibiotic use at the three DFs was restricted to veterinary treatment of diseased animals. Manure at the DFs was stored for up to a year before it was applied to the soil surface.
The three NAGs were Quetico Provincial Park (ON, Canada), the Boundary Waters Canoe Area Wilderness (BWCAW; Minnesota, MN, USA) and the University of Minnesota (Minneapolis, MN, USA). The BWCAW and Quetico Provincial Park are undeveloped wilderness areas situated west of Lake Superior near the Canada–US border comprised of pristine forests and lakes that receive approximately 200 000 and 125 000 recreational visitors each year, respectively (Beymer, 2000; Ontario Parks, 2006).
Soil samples were collected from three different locations within 2 m of each other. Samples (∼500 g) were collected from the soil surface to which manure had been applied in the case of the DFs and from depths of 1–6 inches below the manure treated soil surface at the SFOs except Site SFO1. At Site SFO1, soil samples were collected near (∼20 m) the pen from the edge of an adjacent corn field where there was no visible manure accumulation. Soil samples at the NAGs were collected from grassland (University of Minnesota) and forested land (BWCAW and Quetico Provincial Park) immediately below the O-horizon. Soil samples were immediately transported to the University of Minnesota and processed within 24 h, except for the BWCAW and Quetico Provincial Park samples, which were processed within 72 h.
Enumeration of resistant soil bacteria
Bacteria were dislodged from soil by vortexing 0.5 g of soil (wet weight) suspended in 9 ml of phosphate-buffered saline (PBS, 10 mM, pH 7). Samples were then serially diluted 10-fold in PBS and applied to agar plates using standard spread plating techniques. Agar plates were amended with either 20 mg l−1 of chlortetracycline, 20 mg l−1 tylosin, or 40 mg l−1 of carbadox to enumerate resistant bacteria. Antibiotic resistance levels were calculated as the ratio of bacteria growing on plates supplemented with antibiotics compared to the number of bacteria growing on plates without antibiotics. All initial plate counts were done in triplicate and averaged before calculating resistance levels for each soil sample. The average resistance levels for each of three soil samples collected at a site were subsequently used to calculate mean and s.d. of resistance levels at a site.
Two different nutrient media were used to enumerate bacteria. PYT80 agar plates contained 80 mg l−1 (each) of peptone, tryptone and yeast extract as well as 400 mg l−1 delvocid salt (DSM Food Specialties, Netherlands) and 15 g l−1 granulated agar. Delvocid salt was added as a fungicide; it contained 50% natamycin (w/w). Luria–Bertani (LB) growth medium was composed of 10 g l−1 tryptone, 5 g l−1 yeast extract, 4.9 g l−1 NaCl, 200 mg l−1 delvocid salt and 15 g l−1 granulated agar. PYT80 plates were incubated at 25°C until bacterial colonies were visible (3–5 days); LB plates were incubated for 1–2 days at 37°C. All growth media were sterilized by autoclaving (20 min; 121°C; 15 psig).
Identification of resistant bacterial isolates
Antibiotic-resistant colonies were randomly picked from plates containing 30–300 colonies. Bacteria collected from plates were regrown in liquid culture or collected directly from agar plates. Cells were suspended in 0.5 ml of lysis buffer (5% sodium dodecyl sulfate, 120 mM sodium phosphate, pH 8) followed by three freeze–thaw cycles and a 90 min incubation at 70°C to lyse the cells. Genomic DNA was then purified using a Fast DNA Spin Kit (MP Biomedicals LLC, Irvine, CA, USA).
Bacterial isolates were initially screened by ribosomal intergenic spacer analysis (RISA) to identify unique isolates. Polymerase chain reaction (PCR) was performed using a PTC 100 thermal cycler (MJ Research Inc., Watertown, MA, USA). The ribosomal intergenic spacer region was amplified using primers 1522F (5′-TGCGGCTGGATCCCCTCCTT-3′) and 132R (5′-CCGGGTTTCCCCATTCGG-3′) (Ranjard et al., 2001). The final 50 μl reaction mixture contained: 1 × PCR buffer (Promega, Madison, WI, USA), 4 nmol deoxynucleoside triphosphates, 25 pmol forward and reverse primers, 1.25 units of Taq polymerase (Promega) and ∼1 ng of template DNA. The PCR protocol included a 5 min initial denaturation at 95°C, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min and a final extension for 10 min at 72°C. Amplified products were resolved by electrophoresis on 2% (wt/vol) agarose gels stained with ethidium bromide.
PCR was used to amplify 16S rRNA gene fragments of isolates having distinct RISA patterns using primer set 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1522R (5′-AAGGAGGTGATCCANCCRCA-3′) or primer set 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 907R (5′-CCGTCAATTCCTTTRAGTTT-3′) or primer set 338F and 518R (5′-ATTACCGCGGCTGCTGCTGG-3′) (Edwards et al., 1989; Lane, 1991; Muyzer et al., 1993). Different primer sets were used because some primer sets would generate either no PCR products or multiple products of different sizes. PCR mixtures and conditions were as described above. PCR products were purified using a GeneClean kit (MP Biomedicals) and sequenced using 338F and 907R or 338F and 518R as sequencing primers. Sequencing was performed at the Advanced Genetic Analysis Center at the University of Minnesota using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Multiplex PCR
Chlortetracycline-resistant isolates were examined by multiplex PCR for the presence of 14 different genes encoding for resistance to tetracycline (Ng et al., 2001). These genes confer resistance to tetracycline by three different mechanisms: efflux (tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(K), tet(L), tetA(P)); ribosomal protection (tet(M), tet(O), tet(S), tet(Q)); enzymatic degradation (tet(X)) (Chopra and Roberts, 2001). Multiplex PCR was performed in four separate groups (Group I: tet(B), tet(C), and tet(D); Group II: tet(A), tet(E), and tet(G); Group III: tet(K), tet(L), tet(M), tet(O) and tet(S); and Group IV: tetA(P), tet(Q) and tet(X)). PCR conditions included a 5 min initial denaturation at 94°C, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1.5 min and a final extension for 10 min at 72°C. PCR products were separated on 2% (wt/vol) agarose gels stained with ethidium bromide. Isolates putatively containing genes encoding for tetracycline resistance were identified by comparison with positive controls (Ng et al., 2001) that were simultaneously resolved on 2% (w/vol) agarose gels. Multiplex PCR product specificity was initially confirmed by performing PCR targeting a single gene encoding resistance to tetracycline. About 10% of these PCR products targeting a single gene were randomly selected for further confirmation by nucleotide sequence analysis. In addition, specific PCR products were subjected to nucleotide sequence analysis when genes encoding resistance to tetracycline were detected in an organism not previously known to harbor that gene (for example, tet(A) in a Bacillus sp).
Data analysis
Analysis of variance (ANOVA) was performed to determine the significance of the differences between antibiotic resistance levels at the 10 sites. Differences in chlortetracycline resistance levels at the sites were further examined by pairwise comparison of resistance levels using Tukey honest significant difference. This method used a stringent Type I error rate and used the Studentized range distribution to construct simultaneous confidence intervals for differences of all pairs of means. These statistical analyses were performed using MacAnova software (Version of 02/05/03 Win32s (BCPP5.0), Department of Applied Statistics, University of Minnesota, MN, USA (http://www.stat.umn.edu/macanova).
Principal component analysis (PCA) was performed to evaluate differences between the 10 sites based on the presence or absence of the types of chlortetracycline-resistant bacteria (based on 16S rRNA gene sequences) and their relative abundance. Bacteria having different RISA patterns but at least 98% partial (∼550 bp) 16S rRNA gene sequence identity were grouped together as a single type and their abundance was measured as the percentage of the total number of chlortetracycline-resistant isolates from a particular site on a particular growth medium. PCA was performed using NTSYSpc version 2.11S (Applied Biostatistics Inc., Setauket, NY, USA).
The 16S rRNA gene fragment sequences were compared with sequences obtained from the GenBank database using the BLASTn program (Benson et al., 1999) to determine the phylogenetic affiliation of the isolate. Nucleotide sequences (partial 16S rRNA genes and genes encoding tetracycline resistance) have been deposited in the GenBank database under accession numbers DQ910178-DQ910203, DQ910205-DQ910237, DQ910239-DQ910317 and EF055265-EF055292.
Results
Enumeration of resistant bacteria in soil
Bacteria resistant to tylosin and carbodox were enumerated on nutrient-poor (PYT80) agar plates from soils at nine different sites (Table 2). Mean resistance levels were highly variable for both of these compounds, ranging between 3–55% and 10–47%, respectively, of the plate count values on unamended growth media. However, ANOVA revealed that there were no statistically significant differences between the 10 sites based on tylosin (P>0.15) and carbodox (P>0.20) resistance levels.
Bacteria resistant to chlortetracycline were cultivated on both nutrient rich and nutrient poor growth media from all 10 sites (Table 2), with the exception of Site NAG2 where no chlortetracycline-resistant bacteria could be enumerated on nutrient-rich media. In contrast to the tylosin and carbodox resistance levels, ANOVA suggested that there were statistically significant differences between bacterial resistance to chlortetracycline among the sites, as enumerated on both nutrient-rich (LB; P<0.0005) and nutrient-poor (P<0.05) agar plates. More rigorous statistical analysis (Tukey's honest significant difference; P<0.05) suggested that only the enumerations on nutrient-rich media from sites SFO1 and SFO4 were significantly higher than the other sites. This same test suggested that there were no statistically significant differences among the bacterial resistance levels enumerated on nutrient-poor media.
Identification of chlortetracycline-resistant bacteria
Because differences were detected among the resistance levels to chlortetracycline, more than 750 bacterial strains were isolated from the 10 sites and analyzed in further detail. Bacterial isolates were initially screened by RISA to identify unique strains. Although about 10% of the isolates failed to generate a fingerprint, RISA was able to identify successfully unique bacterial strains among the rest of the isolates. All of the unique strains, as well as the ones that failed to generate a RISA fingerprint, were then identified by partial 16S rRNA gene sequence analysis (Tables 3 and 4).
The type of growth media had a substantial impact on the type of chlortetracycline-resistant bacteria isolated from the different sites. Low G+C Gram-positive (Firmicutes) and high G+C Gram-positive (Actinobacteria) bacteria were the most prominent chlortetracycline-resistant bacteria isolated on nutrient-rich media, although a few members of the Proteobacteria were also detected (Table 3). Particularly common among these isolates were Streptomyces spp., which were the most abundant chlortetracycline-resistant organisms at two of the swine farms (Sites SFO2 and SFO3), two of the DFs (Sites DF2 and DF3) and two of the non-agricultural sites (Sites NAG1 and NAG3). Site SFO1 had the highest diversity of resistant bacteria (16 of the 47 isolates were unique) with a high prevalence of Firmicutes and Proteobacteria and an absence of Streptomyces spp.
The types of chlortetracycline-resistant bacteria enumerated on nutrient-poor growth media were predominantly Chryseobacteria spp. and different types of Proteobacteria, with a bias towards Variovorax spp. (Table 4). Site NAG1 was distinctive compared to the other sites because only Streptomyces spp. were detected on nutrient-poor media. Site SFO1 again contained the most diverse group of resistant bacteria enumerated on nutrient-poor media (11 of 37 colonies were unique).
PCA was performed separately on chlortetracycline-resistant isolates on nutrient-rich and nutrient-poor media to evaluate differences between sites based on the types of resistant bacteria and their abundance (Figure 1). Site SFO1 strongly separated from all the other sites on the first principal component axis for both media types. Site SFO4 separated on the second principal component axis with only the nutrient-rich media (Figure 1a). Similarly, Site NAG2 separated from the rest on the second principal component axis with the nutrient-poor media (Figure 1b).
Principal component analysis of the types (based on partial 16S rRNA gene sequence) and abundance of chlortetracycline-resistant isolates from the 10 sites on (a) nutrient-rich and (b) nutrient-poor growth media. For the nutrient-rich medium, principal component 1 and principal component 2 explain 34 and 22% of variation in the data, respectively. For the nutrient-poor medium, principal component 1 and principal component 2 explain 32 and 17% of variation in the data, respectively.
Multiplex PCR of genes encoding for resistance to tetracycline
All of the chlortetracycline-resistant bacterial isolates (that is, even those identified as redundant by RISA) were characterized by the presence or absence of 14 different genes that encode for tetracycline resistance. Numerous genes encoding for tetracycline resistance were detected, predominantly among the Firmicutes and Proteobacteria that had been isolated from different sites on nutrient-rich media (Table 3). The highest frequencies of detection of these genes encoding for tetracycline resistance were found in bacteria isolated from Sites SFO1 (43 of 47 isolates, with some of them harboring up to three different genes) and SFO4 (26 of 49 isolates). Genes encoding for tetracycline resistance were also commonly detected among bacteria isolated from Site SFO1 on nutrient-poor media (28 of 37 isolates) (Table 4). In contrast, bacteria isolated from the other nine sites that were isolated on nutrient-poor media rarely (2 of 293 isolates) contained one of these 14 genes encoding for tetracycline resistance.
Persistence of chlortetracycline resistance at Sites SFO1 and SFO4
Coincidental to the onset of our study, farming operations ceased at Site SFO1, allowing us to study the long-term persistence of antibiotic resistance. Bacterial enumerations on both nutrient-rich and nutrient-poor media suggested that resistance levels were sustained for at least 18 months and there was no significant temporal variation in resistance levels (Figure 2a). Furthermore, soil samples collected in July 2005 at three different locations at Site SFO1 (at distances of 5, 20 and 100 m from the animal pen) suggested that the former location of the animal pen remained a hot spot for chlortetracycline resistance (Figure 2b).
(a) Chlortetracycline resistance levels at SFO1 between November 2003 and July 2005. Animal feeding operations ceased in November 2003. (b) Chlortetracycline resistance levels as measured in July 2005 at SFO1 at varying distances from the location of the animal pen. Solid bars represent the percentage of chlortetracycline-resistant bacteria enumerated on nutrient-rich medium; open bar represent the percentage of chlortetracycline-resistant bacteria enumerated on nutrient-poor medium.
Approximately 300 chlortetracycline-resistant bacteria were isolated and characterized from soil samples collected from three different locations at Site SFO1 in July 2005. Of the bacteria isolated on nutrient-rich media, there was considerable overlap in the types of resistant bacteria detected at 5 m from the animal pen compared to samples collected 20 m from the pen (Table 5). These bacteria were phylogenetically similar to the bacteria that were previously detected (Table 3), although there was an increase in the frequency of Bacillus spp., which correlated to a decrease in the diversity of resistant bacteria. In contrast, Streptomyces spp. were most prevalent (21 of 41 isolates) among the chlortetracycline-resistant bacteria isolated from the soil collected about 100 m from the animal pen.
Of the bacteria isolated on nutrient-poor media, the bacteria detected at 5 and 20 m from the animal pen in July 2005 (Table 6) were phylogenetically similar to those bacteria previously isolated on nutrient-poor media (Table 4). Further away from the animal pen (100 m), Variovorax spp. were the most commonly detected chlortetracycline-resistant bacteria (18 of 24 isolates). There was also a shift in the prevalence of genes encoding for tetracycline resistance as a function of distance from the animal pen. More than half of the bacteria (31 of 45 isolates) isolated 5 m from the animal pen possessed at least one of 14 genes encoding for tetracycline resistance. In contrast, only a small fraction (4 of 24 isolates) of the bacteria isolated 100 m from the animal pen harbored one of the 14 genes encoding for tetracycline resistance.
Although marginally elevated chlortetracycline resistance levels were originally detected at Site SFO4, we speculated that this result was biased because these soil samples were collected only 1 week subsequent to manure application. Subsequent samples were therefore collected after an additional 10 months of time. Chlortetracycline-resistant bacteria were not detected on the nutrient-rich media, while resistance levels on the nutrient-poor media were statistically similar to all of the other sites (except Site SFO1) (data not shown).
Discussion and conclusions
The subtherapeutic use of antibiotics in agriculture has been a controversial subject for decades (Levy, 1978). Along with rising concerns about the failure of many antibiotics in treating diseases, evidence suggests that the use of agricultural antibiotics is contributing to the spread of antibiotic resistance in the environment (Ferber, 2003; Onan and LaPara, 2003; Wegener, 2003; Rooklidge, 2004). There is also speculation that agricultural use is responsible for continued prevalence of resistance to certain antibiotics after discontinuing their clinical use (Livermore, 2003). However, the lack of causal linkage between agricultural use and the proliferation of antibiotic resistance among pathogenic bacteria has led some people to believe that the economic benefit of antibiotic use in agriculture is sufficient to continue this practice (Phillips et al., 2004).
In our study, we quantified and characterized antibiotic-resistant bacteria in soils at farms using antibiotics for non-therapeutic purposes as well as at farms using antibiotics predominantly for veterinary purposes. The goal of this study was to discern the impact of antibiotic use on the occurrence and abundance of cultivable antibiotic-resistant bacteria in nearby soils exposed to animal waste. Elevated levels of chlortetracycline resistance were quantified in the soils at some, but not all, of the farms that used antibiotics for non-veterinary purposes. When elevated levels of antibiotic-resistant bacteria were enumerated, significant shifts were also observed in the types of chlortetracycline-resistant bacteria as well as the types of genes that encode resistance within these organisms.
Contrary to our expectations, the farm with the highest chlortetracycline resistance levels was the smallest swine farm (Site SFO1), which housed no more than 50 animals at a time. These elevated resistance levels were concomitantly associated with a higher frequency of 14 genes encoding for resistance to tetracycline. We speculate that the excessive exposure to animal manure was responsible for the observed result. While the other farms in our study collected and applied manure to soil at agronomically appropriate rates, Site SFO1 allowed manure to spill uncontrolled out of the animal pen. Previous studies have shown that manure from animals fed subtherapeutic concentrations of antibiotics contains numerous resistant bacteria (Levy, 1978; Aarestrup et al., 2002; Sørum et al., 2006), but that resistance among soil bacteria returned to preapplication levels within 6 months of manure application (Sengelov et al., 2003). We hypothesize, however, that if an excessive quantity of these bacteria is applied to soil, then antibiotic resistance can spread among the indigenous soil bacteria because of lateral gene transfer of the resistance determinants (Andrews et al., 2004). The spread of genes encoding for resistance to indigenous soil bacteria is potentially pertinent because these organisms should be better adapted for survival in the soil, thus potentially serving as a long-term reservoir of antibiotic resistance.
More detailed analysis of the genes encoding tetracycline resistance further suggested that lateral gene transfer was a pertinent factor in the persistence of chlortetracycline resistance at Site SFO1. Numerous bacteria harbored the same gene encoding for tetracycline resistance, similar bacteria (based on 16S rRNA gene sequences) harbored different genes encoding for tetracycline resistance, and individual bacteria harbored multiple types of genes encoding for tetracycline resistance. Particularly noteworthy is the detection of tet(A) in Bacillus spp. and tet(L) in different types of Proteobacteria. Both tet(A) and tet(L) genes encode for efflux pumps that are predominantly associated with gram-negative and gram-positive bacteria, respectively (Chopra and Roberts, 2001; Aminov et al., 2002). The detection of tet(X) in a Sphingobacteria spp., which encodes for a tetracycline-degrading mono-oxygenase (Yang et al., 2004), is also interesting because this gene, thus far, has been found only in Bacteroides spp. (Whittle et al., 2001), which are obligate anaerobes, perhaps suggesting that we have found the natural host of this gene. We are currently characterizing this organism in more detail.
The results from our study have important practical implications. Three of the swine farms were able to subtherapeutically use antibiotics without leading to significantly different numbers or types of chlortetracycline-resistant bacteria compared to the dairy farm soils. These swine farms temporarily stored their animal waste in covered, underground storage pits until it was applied to an appropriate surface area of land per Minnesota state regulations (Minnesota Pollution Control Agency, 2000). We conclude, therefore, that a relatively small quantity of manure containing antibiotic-resistant bacteria can be spread on agricultural soils without significant lateral gene transfer of resistance determinants to the indigenous soil bacteria, allowing the resistant bacteria in the manure to slowly decay in the soil without leading to the proliferation of resistance.
An ancillary goal of our research was to compare resistant bacteria from soils at farms using antibiotics with resistant bacteria from soils unaffected by antibiotic use. Because many antibiotics are natural compounds, numerous soil bacteria would be expected to be resistant even if humans had not been mass-producing various antibiotics since the 1940s (Davelos et al., 2004; Guardabassi and Dalsgaard, 2004). It was somewhat surprising, therefore, that the soil bacteria from the three DFs and two of the farms using antibiotics at subtherapeutic concentrations had similar quantities and types of resistant bacteria compared to the soils that had not been affected by agricultural activity. It is particularly interesting that the bacteria isolated from Quetico Provincial Park (that is, the site least affected by human activity) were exclusively Streptomyces spp., which naturally produce many antibiotics and are equipped with various defense mechanisms.
A principal limitation of the present study is that a cultivation-based approach was used to quantify and characterize chlortetracycline-resistant bacteria in our soil samples. Cultivation-based approaches are known to detect only a small fraction (usually about 1% of soil bacteria) of the total bacterial community (Amann et al., 1995). Although two different growth media were used for chlortetracycline to help limit this bias, we assume that our approach was able to detect only a small fraction of the chlortetracycline-resistant bacteria in these soils. In contrast, cultivation-independent analysis of chlortetracycline resistance would have its own limitations and biases. Cultivation-independent community analysis of chlortetracycline resistance would require that the genes encoding for resistance be analyzed directly (for example, by quantitative PCR), which would have provided a dataset of limited usefulness because the organisms that would have harbored these genes would have remained unknown. Second, cultivation-independent analysis of chlortetracycline resistance would be especially complex because there are now more than 40 different genes that are known to encode for tetracycline resistance (Roberts, 2005).
The diversity and complexity of the genetics of tetracycline resistance also represents another substantial limitation of the present study. Although the multiplex PCR approach used herein was able to reliably detect 14 common genes that encode for resistance to tetracycline, we were unable to detect any gene that encoded for resistance to tetracycline in the majority of our isolates – particularly among the strains isolated from soil samples other than Site SFO1. These bacteria, which did not harbor any of the genes for which we assayed, could have been intrinsically resistant to tetracycline (especially the Streptomyces spp.), harbored one of the other previously discovered genes that encodes for resistance to tetracycline, or harbored a novel gene encoding for resistance to tetracycline. Because soil bacterial communities are particularly diverse (Torsvik et al., 1990), they can also potentially serve as a critical reservoir of genetic material for antibiotic resistance (D’Costa et al., 2006). Additional research is needed to further elucidate the diversity of resistance determinants in environmental hotspots of antibiotic resistance.
Our hypothesis is that the proliferation of antibiotic resistance is principally modulated by the number of antibiotic-resistant bacteria, the genes harbored by these bacteria, the relative ease by which genes encoding for resistance can be laterally transferred among different bacterial strains, and any selective pressures that provide a competitive advantage for antibiotic-resistant bacteria. At the present time, the scientific community appears to be particularly focused on the latter factor (selective pressures), which are primarily linked to antibiotic use. Although we agree that antibiotic use is the most pertinent factor in the global spread of resistance, our research demonstrates that the proliferation of antibiotic resistance is not intrinsically linked to antibiotic use and that the other three factors can also be critically important. We recommend, therefore, substantially more stringent control of fecal waste (both of animal and of human origin) as a viable approach to slow the proliferation of antibiotic-resistant bacteria.
In conclusion, our results suggest that the subtherapeutic use of antibiotics can lead to the proliferation of antibiotic resistance among soil bacteria if an excessive quantity of manure is applied to an insufficient surface area of land. Additional research is needed, however, to confirm or refute this conclusion. In contrast, our results revealed no increase in the quantity of resistant bacteria when animal manure was applied at agronomically acceptable rates. Of particular importance is that the proliferation of antibiotic resistance appears to be associated with specific genes that confer antibiotic resistance. Because these genes were detected among numerous different bacterial populations, lateral genetic exchange appears to be a pertinent mechanism by which resistance develops among soil bacteria.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Aarestrup FM, Hasman H, Jensen LB, Moreno M, Herrero IA, Dominguez L et al. (2002). Antimicrobial resistance among enterococci from pigs in three European countries. Appl Environ Microbiol 68: 4127–4129.
Aarestrup FM, Seyfarth AM, Emborg HD, Pedersen K, Hendriksen RS, Bager F . (2001). Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother 45: 2054–2059.
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA et al. (2002). Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl Environ Microbiol 68: 1786–1793.
Andrews RE, Johnson WS, Guard AR, Marvin JD . (2004). Survival of Enterococci and Tn916-like conjugative transposons in soil. Can J Microbiol 50: 957–966.
Arnold S, Gassner B, Giger T, Zwahlen R . (2004). Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol Drug Safety 13: 323–331.
Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF, Rapp BA et al. (1999). GenBank. Nucleic Acids Res 27: 12–17.
Beymer R . (2000). Boundary Waters Canoe Area – Western Region, 6th edn. Wilderness Press: Berkeley, CA.
Chee-Sanford JC, Aminov RI, Krapac IJ, Garrigues-Jeanjean N, Mackie RI . (2001). Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl Environ Microbiol 67: 1494–1502.
Chopra I, Roberts M . (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65: 232–260.
D’Costa VM, McGrann KM, Hughes DW, Wright GD . (2006). Sampling the antibiotic resistome. Science 311: 374–377.
Davelos AL, Kinkel LL, Samac DA . (2004). Spatial variation in frequency and intensity of antibiotic interactions among Streptomycetes from prairie soil. Appl Environ Microbiol 70: 1051–1058.
De Liguoro M, Cibin V, Capolongo F, Halling-Sorensen B, Montesissa C . (2003). Use of oxytetracycline and tylosin in intensive calf farming: evaluation of transfer to manure and soil. Chemosphere 52: 203–212.
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC . (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17: 7843–7853.
Ferber D . (2003). Antibiotic resistance – WHO advises kicking the livestock antibiotic habit. Science 301: 1027.
Guardabassi L, Dalsgaard A . (2004). Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol 70: 984–990.
Hayes JR, English LL, Carr LE, Wagner DD, Joseph SW . (2004). Multiple-antibiotic resistance of Enterococcus spp. isolated from commercial poultry production environments. Appl Environ Microbiol 70: 6005–6011.
Jackson CR, Fedorka-Cray PJ, Barrett JB, Ladely SR . (2004). Effects of tylosin use on erythromycin resistance in Enterococci isolated from swine. Appl Environ Microbiol 70: 4205–4210.
Lane DJ . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons: New York, pp 115–175.
Levy SB . (1978). Emergence of antibiotic-resistant bacteria in the intestinal flora of farm inhabitants. J Infect Dis 137: 689–690.
Levy SB, Marshall B . (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10: S122–S129.
Lipsitch M, Singer RS, Levin BR . (2002). Antibiotics in agriculture: when is it time to close the barn door? Proc Natl Acad Sci USA 99: 5752–5754.
Livermore DM . (2003). Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis 36: S11–S23.
Minnesota Pollution Control Agency. (2000). Revised feedlot rules at a glance, Minnesota Rules Chapter 7020. Minnesota Pollution Control Agency, St Paul, MN.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Ng L-K, Martin I, Alfa M, Mulvey M . (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes 15: 209–215.
Onan LJ, LaPara TM . (2003). Tylosin-resistant bacteria cultivated from agricultural soil. FEMS Microbiol Lett 220: 15–20.
Ontario Parks. (2006). Quetico Provincial Park management plan review terms of service. www.ontarioparks.com.
Phillips I, Casewell M, Cox T, De Groot B, Friis C, Jones R et al. (2004). Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 53: 28–52.
Ranjard L, Poly F, Lata JC, Mougel C, Thioulouse J, Nazaret S . (2001). Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl Environ Microbiol 67: 4479–4487.
Roberts MC . (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245: 195–203.
Rooklidge SJ . (2004). Environmental antimicrobial contamination from terraccumulation and diffuse pollution pathways. Sci Tot Environ 325: 1–13.
Sengelov G, Agerso Y, Halling-Sorensen B, Baloda SB, Andersen JS, Jensen LB . (2003). Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ Int 28: 587–595.
Sørum M, Johnsen PJ, Aasnes B, Rosvoll T, Kruse H, Sundsfjord A et al. (2006). Prevalence, persistence, and molecular characterization of glycopeptide-resistant Enterococci in Norwegian poultry and poultry farmers 3 to 8 years after the ban on avoparcin. Appl Environ Microbiol 72: 516–521.
Torsvik V, Goksøyr J, Daee FL . (1990). High diversity in DNA of soil bacteria. Appl Environ Microbiol 56: 782–787.
United States Environment Protection Agency. (2004). Risk assessment evaluation for concentrated animal feeding operations EPA 600/R-04/042. National Environmental Publications Internet Site.
Wegener HC . (2003). Antibiotics in animal feed and their role in resistance development. Curr Opin Microbiol 6: 439–445.
Whittle G, Hund BD, Shoemaker NB, Salyers AA . (2001). Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposons CTnDOT. Appl Environ Microbiol 67: 3488–3495.
Yang W, Moore IF, Koteva KP, Bareich DC, Hughes DW, Wright GD . (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 279: 52346–52352.
Acknowledgements
This work was financially supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education and Extension Service (Grant number 2003–35107–13830).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghosh, S., LaPara, T. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J 1, 191–203 (2007). https://doi.org/10.1038/ismej.2007.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.31
Keywords
This article is cited by
-
Clonal relationship, virulence genes, and antimicrobial resistance of Morganella morganii isolated from community-acquired infections and hospitalized patients: a neglected opportunistic pathogen
International Microbiology (2023)
-
Antibiotic resistome mostly relates to bacterial taxonomy along a suburban transmission chain
Frontiers of Environmental Science & Engineering (2022)
-
The influence of antibiotics on the anammox process — a review
Environmental Science and Pollution Research (2022)
-
The combined toxicity of binary mixtures of antibiotics against the cyanobacterium Microcystis is dose-dependent: insight from a theoretical nonlinear combined toxicity assessment method
Environmental Science and Pollution Research (2022)
-
Distribution of antibiotic-resistant bacteria in aerobic composting of swine manure with different antibiotics
Environmental Sciences Europe (2021)