Abstract
Whale-falls represent localized areas of extreme organic enrichment in an otherwise oligotrophic deep-sea environment. Anaerobic remineralization within these habitats is typically portrayed as sulfidogenic; however, we demonstrate that these systems are also favorable for diverse methane-producing archaeal assemblages, representing up to 40% of total cell counts. Chemical analyses revealed elevated methane and depleted sulfate concentrations in sediments under the whale-fall, as compared to surrounding sediments. Carbon was enriched (up to 3.5%) in whale-fall sediments, as well as the surrounding sea floor to at least 10 m, forming a ‘bulls eye’ of elevated carbon. The diversity of sedimentary archaea associated with the 2893 m whale-fall in Monterey Canyon (California) varied both spatially and temporally. 16S rRNA diversity, determined by both sequencing and terminal restriction fragment length polymorphism analysis, as well as quantitative PCR of the methyl-coenzyme M reductase gene, revealed that methanogens, including members of the Methanomicrobiales and Methanosarcinales, were the dominant archaea (up to 98%) in sediments immediately beneath the whale-fall. Temporal changes in this archaeal community included the early establishment of methylotrophic methanogens followed by development of methanogens thought to be hydrogenotrophic, as well as members related to the newly described methanotrophic lineage, ANME-3. In comparison, archaeal assemblages in ‘reference’ sediments collected 10 m from the whale-fall primarily consisted of Crenarchaeota affiliated with marine group I and marine benthic group B. Overall, these results indicate that whale-falls can favor the establishment of metabolically and phylogenetically diverse methanogen assemblages, resulting in an active near-seafloor methane cycle in the deep sea.
Similar content being viewed by others
Introduction
The deep sea, fed by a trickling input of photosynthetically derived carbon, has historically been considered a low-energy, oligotrophic environment. In localized areas, however, oases of elevated microbial biomass and activity do exist (Jørgensen and Boetius, 2007). Perhaps the most well known are hydrothermal vents, emerging along spreading centers and subduction zones, fueled by hot reduced fluids recirculated within the Earth's crust. Although less publicized, equally rich areas of stimulated biomass production and activity also occur in the permanently cold depths of the deep sea, fueled by methane seepage or large organic accumulations (that is, food falls). For example, carcasses of dead whales deliver ∼20–50 ton of organic material to a typically food-limited sea floor, at a rate that is estimated to be 2000 times faster than the supply of planktonic marine snow (Van Dover, 2000; Smith and Baco, 2003). The microbial ecology within these locally active deep-sea habitats has the potential to be unique, particularly with regard to pathways for carbon degradation and nutrient regeneration. This is especially significant when one considers that over half of the sediments covering the deep ocean floor are thought to contain at least 30% skeletal remains of marine animals (Shepard, 1973; Kennett, 1982).
The anaerobic degradation of organic matter is complex, involving many microbial guilds. In particular, methanogenesis and sulfate reduction are the terminal steps in the diagenesis of organic carbon (Jørgensen, 1982). These processes are thought to be mutually exclusive in most environmental settings and controlled, in large part, by the amount of available sulfate (Reeburgh and Heggie, 1977; Winfrey and Zeikus, 1977). Archaeal methanogenesis is typically considered the dominant process in low-sulfate habitats, such as freshwater lakes, whereas bacterial sulfate reduction dominates in the sulfate-replete marine environment. Sulfate-reducing bacteria outcompete methanogenic archaea for common substrates, including hydrogen and acetate; thus, it is thought that sulfate reduction generally precludes methanogenesis as the primary terminal step in the microbial diagenesis of organic carbon in marine sediments (Kristjansson et al., 1982; Lovley et al., 1982).
The quantity and quality of organic carbon in sediments can also be a major factor determining the distribution and metabolism of microbes (Capone et al., 1983). In certain anoxic marine environments where input of organic carbon is high, including mangroves, eel grass beds, estuaries, swamps and polluted intertidal sediments, both sulfate reduction and methanogenesis can occur simultaneously (Oremland and Taylor, 1978; Oremland et al., 1982; Senior et al., 1982). Similarly, in hypersaline environments, the use of non-competitive methylated substrates by methanogens also enables co-habitation by both functional guilds (Visscher et al., 2003). In the marine environment, methanogenesis and sulfate reduction are often temporally or spatially separated (Cappenberg, 1974; Mountfort and Asher, 1979); for example, methanogenesis is typically dominant in deeper sulfate-depleted sediment horizons below the zone of active sulfate reduction. The methanogenic conversion of organic carbon to CO2 and CH4 below the depth of sulfate depletion persists at very low rates and is thought to account for only a small fraction of the carbon catabolism observed in these marine sediments (Senior et al., 1982). A high surplus of biologically available organic carbon, however, may have a significant influence on the overall habitat distribution of methanogenic archaea and their contribution to carbon breakdown in these environments.
Investigating the diversity and distribution of natural microbial communities in the permanently low-temperature sediments of the deep sea is critical to our understanding of global biogeochemical cycles, especially with regard to the breakdown and recycling of organic carbon. A unique opportunity in which to study the diagenesis of organic matter and the impact of high organic loading on deep-sea microbial assemblages arose with the serendipitous discovery of a decomposing whale at 2893 m depth in Monterey Bay, California (Goffredi et al., 2004). In this study, we combine molecular analyses with geochemical measurements in the sediments underlying this deep-sea whale-fall to characterize changes in the magnitude and distribution of the archaeal community, compared to the surrounding deep-sea sediments. We assessed whether microbial communities were enriched under the whale-fall, if they changed over time, and the extent and impact of this enrichment on the surrounding sea floor. Regular field sampling was conducted to examine the temporal changes in the whale-fall-associated microbial assemblages, an ecological task that is typically difficult to accomplish for deep-sea environments.
Materials and methods
Site description and sampling
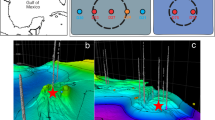
Sediment samples were obtained from a whale-fall in Monterey Canyon, California (Figure 1). The whale-fall was originally discovered in February 2002 (Goffredi et al., 2004; 36.78°N/122.08°W, 2893 m depth; Figure 1a, star symbol). Samples were collected by means of the remotely operated vehicle Tiburon (owned and operated by the Monterey Bay Aquarium Research Institute). Sampling occurred over a period of 59 months (from October 2002 to January 2007) following the discovery of the 2893 m whale-fall (Table 1 and Figure 1); 8 months––dive T486; 33 months––dive T769 (Figures 1b and c); 45 months––dive T917 (Figure 1e); 51 months––dive T991 (Figure 1f); and 59 months––dive T1069 (not shown). Chemical data are shown for two other whale-falls artificially implanted at 385 m (36.79°N/121.89°W) and 1018 m (36.78°N/122.08°W) depth in Monterey Canyon in April 2005 and October 2004, respectively (Braby et al., 2007).
(a) Shaded relief map of the continental margin off Monterey Bay showing the whale-fall location at 2891 m depth (large white star). Modified from Goffredi et al., 2004. (b, c, e, f) Photo mosaics of digital still images. (d) Schematic of the whale skeleton at 2893 m showing areas of sediment collections and sampling strategy. (b, c) At 33 months (dive no. T769, November 2004), (e) 45 months (dive no. T917, November 2005) and (f) 51 months (dive no. T991, May 2006).
Sediments were sampled directly beneath the whale-fall (0 m distance away), and at 1, 3 and 10 m distance from the center of the carcass (the latter considered to be ‘reference’ sediment, initially outside the influence of the whale-fall; Figure 1d). Changes in archaeal community composition were monitored both with increasing distance from the whale-fall and with increasing depth within the sediment column. Spatial sampling, thus, included these two different parameters (Table 1): distance from the whale-fall and depth within the sediment (referred to as ‘depth horizon’).
Sediment samples were collected with push cores and stored at 4 °C until they were processed (usually shipboard within 0–5 h) and subdivided for chemical analysis or molecular studies. From the push core sleeve, samples were extruded upwards in 3-cm-thick sections and subsampled using cutoff 1 and 5 ml syringes. Samples were then subdivided for cell counts via fluorescent in situ hybridization (FISH) (∼0.5 g sample wet weight in 2–4% paraformaldehdye as described below), nucleic acid analyses (approximately 15 g of sediment frozen at −80 °C) and pore-water chemistry (6–10 g for CH4 and ∼15 g sediment for SO4 and H2S, as described below). Additional samples were preserved in ethanol or flash-frozen in liquid nitrogen.
Sediment chemistry
Sediments were collected and analyzed for percent total carbon and nitrogen, stable isotopes (δ13C and δ15N) of bulk carbon and nitrogen, concentration and δ13C of methane specifically, as well as concentrations of sulfide and sulfate. For methane analysis, 6–10 g sediment plugs were added (∼1:1) to 1 M NaOH in gas-tight serum vials. Greater than 99.5% of the methane in the sediment is typically present in the gas phase. Methane concentration in the headspace of the bottles was determined with a Shimadzu mini-2 gas chromatograph equipped with a flame ion detector and magnesium perchlorate trap, with a 2 ml injection loop. A 9.93 p.p.m. methane standard was used for calibration. Sediment porosity was measured to be 40–60%, via drying of sediments at 65 °C for 24 h, and was taken into account in the final methane calculations. Compound specific isotopic values for methane from eight selected samples representing different methanogen assemblages were measured via gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS; Isotech Laboratories Inc., Champaign, IL, USA).
To collect pore waters for both sulfide and sulfate measurements, sediment samples were centrifuged (1380 g for 15 min) in cutoff, stoppered 10 ml syringes without a headspace (Barry et al., 1996). Pore fluid samples were transferred to gas-tight syringes via a needle through the side wall of the 10 ml syringe, minimizing exposure to oxygen, and preserved immediately in either 0.5 M zinc acetate or 0.5 M barium chloride (1:1) for sulfide and sulfate, respectively. Sulfide was measured via the colorimetric cline assay (Cline, 1969). A 200 mM sodium sulfide stock solution was made by dissolving Na2S in N2-sparged distilled water (adjusted to pH 10 with NaOH), and decanted into anaerobic vials. Sulfide standards (15–1000 μM final concentrations) for the Cline assay were made from this stock solution via serial dilution with N2-sparged distilled water. Sulfate in the pore fluids was determined by turbidometry using a spectrophotometer (Association APH, 1976; Gieskes et al., 1991). The turbidity of a given reaction was measured at 420 nm and expressed in units derived from a standard curve prepared by use of barium sulfate suspensions (0–28 mM).
For percent carbon and nitrogen, as well as stable isotopes on bulk sediments, samples were dried at 65°C for 24 h, then milled to a fine powder. Dry material (0.5–2.0 mg) was placed into a tin capsule and combusted in a Eurovector (Milan, Italy) elemental analyzer. The resulting N2 and CO2 gases were separated by gas chromatography and admitted into the inlet of a Micromass (Manchester, UK) isoprime isotope ratio mass spectrometer (IRMS) for determination of 15N/14N and 13C/12C ratios. Typical precision of analyses was ±0.5‰ for δ15N and ±0.2‰ for δ13C where δ=1000(Rsample/Rstandard)−1‰, where R=15N/14N, the standard for δ15N is atmospheric nitrogen and Peedee Belemnite for δ13C. Egg albumin was used as a reference material during each run.
Microscopy
Samples for microscopy were preserved shipboard for 4–12 h at 4 °C in phosphate-buffered 4% paraformaldehyde, rinsed twice with 1 × phosphate-buffered saline (PBS), transferred to an equal volume of PBS/ethanol and stored at −20 °C. Pre-processing for FISH included dilution of sediments 1:20 (vol/vol) in 1 × PBS, containing 0.01 M pyrophosphate (final concentration), to a final volume of 2.0 ml, sonication on ice with a sonicating wand (Branson sonifier 150) for two 10 s pulses at 8 W and shaking at 150 r.p.m. for 15 min at 35 °C. Samples were briefly centrifuged to pellet large particles, and the supernatant was collected onto a 0.2 μm Millipore polycarbonate filter. Catalyzed reporter deposition-FISH (or CARD-FISH) using tyramide signaling and probes labeled with heat-stabilized horseradish peroxidase was performed according to the protocol of Pernthaler et al. (2004). For permeabilization of the target cells, a 40 μl subsample of sonicated sediment was pipetted into 100 ml of 1 × TE (10 mM Tris-HCl, 1 mM EDTA, pH 9.0) in glass dish and microwaved in a histological microwave oven (Microwave Research & Applications Inc., Laurel, MD, USA) at 65 °C for 2 min. Subsequently, the slurry was put onto ice and centrifuged for 5 min at 1000 g at 4 °C. To inactivate endogenous peroxidases, samples were first exposed to 0.1% H2O2 (in 1 × PBS for 1 min at room temperature) and centrifuged at 1000 g for 4 min. Hybridization buffer was made as described previously, using 35% formamide concentration (vol/vol) and 0.5 ng μl−1 of a horseradish peroxidase-labeled oligonucleotide probe (Pernthaler et al., 2004). The universal bacterial probe set EUB338 I-III (Daims et al., 1999) and a universal archaeal probe, Ar915 (Stahl and Amann, 1991), both labeled with horseradish peroxidase (Biomers.net, Ulm, Germany), were used for cell counts. Independent checks for nonspecific hybridization by the CARD-FISH procedure were negative. Hybridizations were conducted at 46 °C for 2–3 h, followed by a 15 min wash in 1 × PBS (at room temperature) and a 10 min amplification step at 37 °C. Amplification buffer contained 0.0015% H2O2 and 0.5 μg of tyramide labeled with Alexa488 (green) or Alexa546 (red) specifically. Cells were post-stained with 4′,6′-diamidino-2-phenylindole (5 μg ml−1) for 1 min and counted by epifluorescence microscopy using a DeltaVision restoration microscopy system (Applied Precision LLC, Issaquah, WA, USA).
Nucleic acid extraction
For 16S rRNA analysis, total nucleic acids were extracted from 3-cm-depth sediment intervals, with each representing a 3-cm-depth interval. Cell lysis and DNA extraction from ∼0.5 g (wet weight) of sediment was conducted using the Ultra-Clean Soil DNA kit (MoBio Laboratories Inc., Carlsbad, CA, USA). The protocol was modified by two initial 5-min incubations at 70 °C followed by beadbeating (using a Bio101 Fastprep machine, speed 5.5 for 45 s; Thermo Electron Corporation, Whaltham, MA, USA). The remainder of the extraction procedure was carried out according to the manufacturer's instructions, with the exception of a second 4 °C incubation in IRS (Inhibitor Removal Solution® (MoBio Laboratories Inc., Carlsbad, CA, USA)) solution between solutions S2 and S3. DNA concentrations were determined using a Nano-Drop 1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA).
PCR, clone library construction and sequencing
SSU rRNA (16S) and methyl-coenzyme M reductase (mcrA) genes were amplified by PCR from purified DNA samples. PCR mixtures (25 μl) contained 0.4 μM of either Archaea-specific 16S rRNA primers, 20F and 958R (Delong, 1992), or mcrA-F and mcrA-R for the mcrA analysis (Luton et al., 2002). Reactions also contained 2.5 μl of 10 × PCR buffer (containing 2 mM MgCl2), 0.2 mM each of deoxynucleotide triphosphates and 0.025 U of Eppendorf HotMaster Taq (Westbury, NY, USA). Thermal cycling conditions included 45 s each of denaturation at 94 °C, annealing at 54 and 50 °C, respectively for 16S rRNA and mcrA, elongation at 72 °C (25–30 cycles) and a final 6 min of elongation at 72 °C. PCR products were cloned using the Topo TA cloning kit (Invitrogen, Carlsbad, CA, USA). Clone libraries of PCR-amplified archaeal 16S rRNA genes were constructed from 2 to 3 depth horizons from each core, with 35–119 clones analyzed for each library, depending on the diversity of recovered ribotypes. A summary of the samples analyzed is included in Table 2. To group similar clone types for subsequent sequence analysis, 16S rRNA clones were subjected to restriction fragment length polymorphism (RFLP) analysis using the restriction enzyme HaeIII (New England Biolabs, Beverly, MA, USA). Inserts from clones chosen for sequencing were amplified with M13 primers, as above except with Taq polymerase from Promega (Madison, WI, USA). M13 amplicons were cleaned prior to sequencing with MultiScreen HTS plates (Millipore Corporation, Bedford, MA, USA). Sequencing reactions were performed on these products using the Genome Lab DTCS Quick Start Kit (Beckman Coulter, Fullerton, CA, USA), then precipitated with glycogen and sodium acetate, resuspended in 40 μl of formamide and run on a CEQ 8800 Genetic Analysis System (Beckman Coulter). Sequences acquired during this study were deposited in GenBank (accession numbers EU084511–EU084526, EU220725 and EU201175–EU201182). Library designations are R (Ruby, 2893 m whale-fall)_no. (months post discovery)_0, 1 or 10 m distance from whale, and s, m or d for shallow (0–3 cm), mid (3–6 cm) or deep (9–12 or 12–15 cm) depth horizons, respectively. Additional sequences were obtained from GenBank and compiled and aligned with our 16S rRNA sequences using ARB Fast Aligner (Ludwig et al., 2004) and Sequencher v4.5 (Gene Codes, Ann Arbor, MI, USA). Phylogenetic trees were generated using PAUP v4.0b10 (Swofford, 1998). The Kimura two-parameter model was used to estimate evolutionary distance. Maximum-parsimony and neighbor-joining analyses were conducted with heuristic searches using 100 and 5000 non-parametric bootstrap replicates, respectively, to assign confidence levels to nodes. mcrA gene sequences were first translated in Sequencher v4.6 and then aligned manually in MacClade v4.08. Total character difference was used to estimate evolutionary distance and a neighbor-joining analysis was conducted using 5000 non-parametric bootstrap replicates, to assign confidence levels to nodes.
Terminal restriction fragment length polymorphism analysis
Terminal RFLP (T-RFLP) was used to further characterize the diversity and relative proportion of methanogen-associated 16S rRNA genes in sediment samples. 16S rRNA genes from purified DNA samples were PCR-amplified using archaeal primers 20F (fluorescently labeled with WellRED dye D4, Sigma-Proligo, St Louis, MO, USA) and 958R using the same PCR conditions as described above for unlabeled PCR reactions. For a few samples that were particularly difficult to amplify, 20 cycles of PCR were performed using unlabeled primers, followed by a subsequent reaction using 1 μl of the unlabeled PCR product for an additional 25 cycles using the fluorescently labeled forward primer. For each sediment sample, duplicate PCR amplifications were performed and pooled prior to digestion with either HaeIII or Sau961 (for 6–8 h at 37 °C; New England Biolabs). Digest reactions contained 3 μl DNA, 0.5 μl of enzyme and 2 μl 10 × buffer diluted with purified H2O to a final volume of 20 μl. HaeIII and Sau961 were chosen based on in silico analysis of known methanogen 16S rRNA sequences using the restriction enzyme cutting program in Sequencher v4.6 (Gene Codes). Digested products were precipitated using glycogen and sodium acetate as carriers (as above for direct sequencing) and resuspended in 40 μl of formamide containing 0.5 μl of an internal dye standard (GenomeLab DNA size 600; Beckman Coulter). Fluorescently labeled fragments were separated by capillary electrophoresis and analyzed on a CEQ 8800 Genetic Analysis System (Beckman Coulter). Fragment sizes were parsed to the nearest two whole base pairs and relative abundances were estimated using the CEQ 8800 Fragment Analysis software. Clones positively identified via sequencing were used as controls for amplification, restriction and sizing during the T-RFLP analysis.
Quantitative PCR analysis of archaeal mcrA genes
The copy number of mcrA genes in each DNA extract was determined using real-time PCR. Primers, modified from Luton et al. (2002), were used to amplify an ∼470-bp fragment from the archaeal mcrA gene (mcrA-f, 5′GGTGGTGTMGGATTCA-CACAR-3′, Tm 56.5 °C, and mcrA-r, 5′-TTCATTGCRTAGTTWGGRTAG-3′, Tm 50.0 °C). Triplicate real-time PCR reactions (20 μl) contained 0.5 mM of each primer and 1 × PCR buffer containing MgCl2, dNTPs and Amplitaq Gold in the SYBR green master mix (Applied Biosystems, Foster City, CA, USA). Purified DNA (1 μl) was added as template (DNA extracts ranged from 10 to 160 ng μl−1 but were normalized to 10 ng μl−1 and diluted 100 ×). All reagents were RNase- and DNase-free. A standard curve was generated using serial dilutions of a pBAD-derived plasmid (Invitrogen) containing a known partial mcrA sequence. This template was CsCl-purified to reduce the presence of inhibitory compounds, and linearized to more closely approximate environmental DNA. The slope of this standard curve (regression lines of Ct versus log N, the log of initial DNA concentration in standard templates) was used to estimate amplification efficiency for these particular primers in our quantitative PCR (QPCR) assays (∼82%, slope of Ct versus concentration=3.86). Reactions were carried out using the Sequence Detector System ABI Prism 7300 (Applied Biosystems). Cycling parameters included an initial denaturation of 95 °C for 3 min, followed by 45 cycles of 95 °C (30 s) and 60 s of annealing/extension at 58.5 °C, determined empirically. A dissociation curve from each QPCR reaction was examined to further ensure proper target sequence amplification.
Results
Chemistry
Sulfate was depleted beneath the whale-fall, relative to both seawater and reference cores (∼28 mM), with the lowest measured value of 9 mM SO4 at a depth horizon of 12–15 cm (Table 1). Sulfide concentrations at the 2893 m whale-fall were generally low (from undetectable levels to only 60 μM; Table 1); however, this trend appears unique to this particular whale-fall as significant sulfide levels were observed at other whale-fall sites in Monterey Canyon (up to 2.5 mM, data not shown).
Methane concentrations were elevated in sediments directly beneath the whale (up to 4230 μmol CH4 g−1 sediment; Table 1) relative to the reference sediments. Two reference cores taken approximately 450 m from the whale-fall (and ∼60 m apart) had methane values of ∼0.2 μM (Bill Ussler, personal communication, Monterey Bay Aquarium Research Institute). Spatial (lateral) heterogeneity in methane levels was recorded at 45 months, with higher values observed in sediments immediately underlying the whale-fall (up to 1455 μmol CH4 g−1 sediment at 6 cm depth, closed circles in Figure 2a), compared to a core (PC47) taken at a distance of 1 m away (129 μmol CH4 g−1 sediment, closed diamonds; Figure 2a). Associated stable carbon isotope values for methane were measured for a subset of samples beneath, or adjacent to, the whale-fall. The greatest difference in δ13CH4 was observed between different depth horizons (3–6 cm versus 9–12 cm) within a single core (PC47), taken 1 m from the whale-fall, with δ13CH4 ranging from −39.6‰ to −58.6‰ (see 45 months, Table 1; closed diamonds in Figure 2a), while a second core (PC43) appeared to have more constant δ13CH4 values (∼−50‰; Table 1).
Pore-water profile of (a) total CH4 (μmol per g sediment) under the 2893 m whale-fall (0 m, T917_PC43), and at 1 m (T917_PC47) and 10 m (T917_PC52) distance away. δ13CH4 (‰) are noted next to each depth horizon, along with the predominant methanogen detected in 16S rRNA libraries. (b) Total organic carbon (% TOC) for the same sediment samples and depth horizons with δ13C values (‰) noted next to each depth horizon. All samples were taken at 45 months.
High levels of carbon (total organic carbon; TOC) were observed in sediments under the 2893 m whale-fall for all time points investigated. This enrichment in organic carbon was established relatively early, with no noticeable change in surface sediments from 8 (3.4% TOC) to 59 months (3.2% TOC) post-discovery (Table 1). Spatial relationships (vertical and lateral) in TOC were investigated for cores collected directly beneath the whale-fall (0 m distance), and at 3 and 10 m distances from the center of the carcass (Figure 3). At 59 months, the greatest enrichment was observed in shallow depth horizons (0–3 cm), averaging 2.5±1.1 total wt % (n=7), compared to deeper (9–15 cm) sediment depths (1.8±0.5 total wt %, n=8; Figure 3a). There was a significant decrease in total carbon occurring at 3 cm depth (decreasing by 44±12 relative %), indicating a very shallow and localized zone of enriched carbon immediately under the whale-fall (Figure 3a, open squares and circles). A similar sharp decrease in carbon was observed in sediments collected at a distance of 3 m from the whale-fall (Figure 3a, open triangles and diamonds). At 59 months, elevated levels of TOC were also observed for sediments collected 10 m from the whale-fall; however, the decrease in % TOC with sediment depth was much less (decreasing by only 13%; Figure 3a, closed diamonds). For two artificially implanted whale-falls with known dates of origin, we observed slightly less elevated carbon in underlying sediments at 24 and 20 months after deposition on the sea floor (1.4–2.1%; Figures 3b and c).
Total carbon (%) in sediments under the (a) 2893 m whale-fall (0 m), and at 3 and 10 m distances away (samples were taken at 59 months). For comparison, similar measurements are shown for whale-falls at 1018 m (b) and 385 m (c) depth at earlier stages in their decomposition (24 and 20 months post-deposition on the sea floor, respectively).
The lowest TOC value (0.7%) was measured 1 m from the whale-fall (PC47, Table 1) in a sediment horizon with a δ13C value of −22.2‰ (Figure 2b) and heterogeneous δ13CH4. This particular core showed the greatest difference in δ13C TOC among different depth horizons with a δ13C of −18.7‰ in the 0–3 cm horizon and −22.2‰ in the 9–12 cm horizon (Table 1 and Figure 2b). With the exception of PC47, however, bulk δ13C TOC values beneath the whale-fall were not significantly different from the 10 m sediments (−20.2±0.7‰ (n=5) versus −20.3±0.7‰ (n=4), respectively, for all depth horizons at both 33 and 51 months; Table 1, not all data are shown).
Total nitrogen ranged from 0.2% to 0.5% immediately underlying the carcass and from 0.2% to 0.3% for sediments collected at 3 and 10 m distance from the whale-fall. δ15N values were variable and ranged from 7.5‰ to 12.6‰, with no obvious spatial or temporal pattern (Table 1).
Archaeal communities associated with the whale-fall
Cell counts revealed an increase in total microbial abundance (∼1.6 ×). Under the whale-fall, with archaea representing 10–40% of the total cells detected by FISH microscopy. Total microbial counts (bacterial and archaea) were generally higher (1.5–2.2 ×) in shallow (0–3 cm) versus deep depth horizons (9–15 cm), a trend that was consistent in both 0 and 10 m cores. Similarly, a decrease in the yield of genomic DNA with increasing depth in the sediment was also observed, from 4 to 5 μg DNA g−1 sediment recovered from the 0–3 cm depth horizon and 0.5–1 μg DNA g−1 sediment recovered from the 9–15 cm horizon.
At 33 months, a comparison was made between archaeal assemblages in sediment immediately underlying the whale-fall and at a ‘reference’ site 10 m away. The majority of phylotypes recovered from ‘reference’ sediments (T769_PC55) were related to uncultured marine Crenarchaeota, including phylotypes associated with groups previously recovered from deep-sea sediments (Li et al., 1999; Vetriani et al., 1999; Knittel et al., 2005; Inagaki et al., 2006). Specifically, archaeal assemblages in the reference core from the 2893 m whale-fall primarily consisted of marine group I (MGI) and marine benthic group B (MBGB), representing up to 98% and 27% of the libraries from ‘reference’ sediment, respectively (Table 2 and Figure 4). The dominant Crenarchaeota MGI phylotypes in both depth horizons within reference sediments were related to an uncultured archaeon from a fish gut (R33_10d_G7; van der Maarel et al., 1998) as well as an uncultured Crenarchaeote from sediments in the Japan Trench (R33_0s_B10; Li et al., 1999). These MGI phylotypes fell within the MGI-α microcluster, which contains both surface and deep-water taxa (Massana et al., 2000). The dominant Crenarchaeota MBGB phylotype (R33_10d_E11), present only in the deeper 12–15 cm layer of the reference core, was 98% similar to an uncultured archaeon from deep-sea sediments in the Atlantic Ocean (Table 3 and Figure 5; Vetriani et al., 1999). Phylotypes related to the Euryarchaeota were also found only in the 12–15 cm layer (representing up to 24% of the recovered phylotypes; Table 3 and Figure 4), including a phylotype (R33_10d_G1) related to uncultured Euryarchaeota from sediments in Skan Bay, Alaska and other members of the Thermoplasmatales-related marine benthic group D, (Girguis et al., 2003; Kendall et al., 2007). Additional, as yet unclassified Crenarchaeota (for example, R33_0s_F10), related to freshwater and soil archaea (Bintrim et al., 1997; Stein et al., 2002), made up the remainder of the sequence diversity (11%; Table 3 and Figure 4). No methanogen-affiliated sequences were recovered from the 10 m reference core.
Phylogenetic relationships of archaea associated with a 2893 m whale-fall in Monterey Canyon (CA), based on sequence divergence within a 938-bp region of the 16S rRNA gene, to selected cultured and environmental sequences in public databases. Thermotoga maritima (AJ401021) and Aquifex aeolicus (AE000709) were used as outgroups (not shown). Numbers next to nodes correspond to bootstrap values based on neighbor-joining distance (top number, 5000 replicates) and maximum-parsimony (bottom number, 100 replicates) methods (– indicates no bootstrap support available). GenBank accession numbers for sequences obtained in this study are EU084511–EU084526). Taxon designations correspond to those listed in Table 3. For library designations, refer to Table 2.
In contrast, archaeal assemblages directly beneath the whale-fall (T769_PC18), at the same time point (33 months), were distinct from the surrounding communities, with methanogen-affiliated sequences dominating the archaeal diversity. In both shallow and deep sediment horizons, methanogen phylotypes represented between 36% and 98% of the archaeal diversity observed at all time points (Table 2 and Figure 5). Vertical heterogeneity in methanogen abundance was observed at the 33-month time point with Methanogenium-related phylotypes accounting for approximately 31% of the archaeal diversity in the 0–3 cm depth, yet dominating in the deeper 5–8 and 12–15 cm layers (representing up to 98% of the diversity; Table 2 and Figure 4). This trend in vertical methanogen abundance was confirmed by QPCR analysis of the mcrA gene with 30 × greater mcrA copy numbers detected at 12–15 cm compared to 0–3 cm.
Most of the methanogenic phylotypes in sediments underlying the whale-fall (for example, R33_0s_H2) were highly related (99% similarity in 16S rRNA) to Methanogenium marinum from Skan Bay, Alaska (Table 3 and Figure 5; Chong et al., 2002). Additional methanogenic phylotypes included those related to Methanococcoides alaskense (99% similarity, R33_0s_B2), present in both sediment horizons, and Methanosarcina baltica (98% similarity, R33_0s_B1), present only in the deeper 12–15 cm layer (Table 3 and Figure 5; Singh et al., 2005). In the upper sediment horizon (0–3 cm), MGI made up much of the remainder of the recovered phylotpes (53%, R33_0s_B10; Figure 5), including similar MGI phylotypes as recovered from reference sediments (R33_10d_G7; Table 3 and Figure 5). The relative increase in abundance of methanogens under the whale-fall was additionally supported by QPCR analysis of the methyl-coenzyme M reductase (mcrA), with 40–120 × greater mcrA signal in sediments underlying the whale-fall compared to reference sediments (data not shown).
Changes over time
Over a period of 43 months, methanogen-affiliated phylotypes dominated the archaeal diversity directly beneath the whale-fall (n=4 time points; Figure 6). Specific notable changes in methanogen diversity included a transition from dominance of members of the Methanococcoides at 8 months (in the 0–3 cm horizon beneath the whale-fall) to the later establishment of Methanogenium, detected throughout the sediment column, at all subsequent time points (Table 2 and Figure 6). Methanogenium represented 31%, 80% and 85% of the recovered ribotypes in surface sediments (0–3 cm) collected at 33, 45 and 51 months, respectively (Figure 6). Similarly, Methanogenium represented 56–96% of the recovered ribotypes in deeper (9–15 cm) sediment layers (Table 3 and Figure 6). We believe the whale-fall was well into the process of degradation (duration on the sea floor was estimated to be 6–12 months at the time of discovery; Goffredi et al., 2004); thus, characterization of earlier archaeal assemblages at this site was not possible. However, early measurements for a whale-fall at 1018 m depth, although not featured in this study, revealed that at 3 months post-implantation, methanogen-affiliated phylotypes represented only 18% of the total recovered archaeal phylotypes in the shallow 0–3 cm depth horizon and none were detected in the deeper 9–12 cm horizon (data not shown). Similar to the 8-month time point for the 2893 m whale-fall, methanogens present early on at the 1018 m whale-fall, as well as another whale-fall at 1820 m depth, were predominantly methylotrophic, including Methanococcoides and the metabolically versatile Methanosarcina and Methanolobus spp. (3–13 months, data not shown).
Changes over time in the abundance of archaeal subgroups recovered from sediments immediately underlying the 2893 m whale-fall at four different time points (8, 33, 45 and 51 months after discovery). (a) Surface (0–3 cm) depth horizons and (b) deeper depth horizons (12–15 cm for 33 and 51 months and 9–12 cm for 45 months; see also Table 1). Data shown for the 33-month time point are the same as shown in Figure 4, for sediments underlying the whale-fall.
T-RFLP as a diagnostic for methanogenic taxa in marine sediments
Fingerprinting, by T-RFLP analysis, of the archaeal community in surface sediments (0–3 cm) underlying the whale-fall revealed dominant fragments at 118, 246 and 323 bp with Sau961 (Figure 7a) and 220, 243 and 183 bp with HaeIII, which were determined to be affiliated with Methanosarcina, Methanogenium and Methanococcoides, respectively, based on digestion of sequenced 16S rRNA clones from this study and in silico predictions. T-RFLP analysis of archaea resolved the dominant methanogen-affiliated taxa and generally supported the proportions recovered in the clone libraries from the four different time points examined. Percent representation for methanogens fell within 25±13% on average for both methods and confirmed the transition from Methanococcoides to Methanogenium as the dominant methanogen between 8 and 33 months (Figure 7b).
Fingerprinting, by terminal restriction fragment length polymorphism analysis (T-RFLP), of the archaeal community in surface sediments (0–3 cm) underlying the whale-fall. Dominant fragments at (a) 118, 246 and 323 bp with Sau961 (shown) and 220, 243 and 183 bp with HaeIII (not shown) were derived from Methanosarcina, Methanogenium and Methanococcoides, respectively, based on digestion of known clones and in silico calculations. (b) T-RFLP analysis of archaeal 16S rRNA genes resolved the two dominant methanogen taxa (Methanogenium and Methanococcoides) and supported their relative proportions in clone libraries from the various depth horizons at four different time points examined. Clone library percentages are the same as depicted in Figure 6a.
A shift in archaea from methane producers to methane consumers
Lateral heterogeneity in archaeal diversity was also observed at a later time point (45 months) between sediments directly under the whale-fall (T917_PC43) versus those at a distance of 1 m away (T917_PC47, Table 2). Compared to the Methanogenium-dominated archaeal assemblages within methane-producing sediments immediately underlying the whale-fall (T917_PC43), phylotypes affiliated with the uncultured methanotrophic archaeal anaerobic methane-oxidizing euryarcheota group 3 (ANME-3) group were unexpectedly dominant (64% of phylotypes recovered) in the 9–12 cm depth horizon of a core (PC47) collected 1 m away (Table 2). The phylotype R33_1d_H12 was 99% similar to an uncultured ANME-3-related archaeon from anoxic methane seep sediments from the Eel River Basin (Figure 5; Orphan et al., 2001; Niemann et al., 2006). PCR surveys of the methyl-coenzyme M reductase (mcrA) gene confirmed the potential shift from methane-producing to -consuming archaea with the majority of mcrA ribotypes (for example, R45_1d_D1_mcr) recovered from the 1 m core (PC47) belonging to the ‘novel mcr group f’ (ANME-3; Losekann et al., 2007), while the mcrA ribotypes recovered from the 0 m core (PC43) grouped with known methane producers (for example, R45_0d_D5_mcr; Figure 8). Archaeal diversity in the shallower 3–6 cm depth horizon of PC47 was comprised of Methanococcoides (36% of the 16S rRNA phylotypes recovered) in addition to ANME-3 (10%), with crenarchaeotal groups MBGB (36%) and MG1 (9%) making up the balance. The variation in archaeal diversity observed both laterally and vertically on the centimeter scale suggests substantial heterogeneity and complexity in both the microorganisms and processes relating to methane cycling within the whale-fall habitat.
Phylogenetic relationships of archaea associated with a 2893 m whale-fall in Monterey Canyon (CA), based on amino-acid divergence (166 amino acids) within a region of the methyl-coenzyme M reductase (mcrA) gene, to selected cultured and environmental sequences in public databases. Methanopyrus kandleri (AE009439) was used as the outgroup. Numbers next to nodes correspond to bootstrap values based on neighbor-joining distance (5000 replicates). GenBank accession numbers for sequences obtained in this study are EU201175–EU201182. Group ‘f’ designation bases on Losekann et al., 2007.
Discussion
Archaeal communities associated with whale-falls
Time-course investigation of the microbial community within an organically enriched whale-fall habitat in Monterey Bay, CA, revealed a previously underappreciated niche for methanogenic and methanotrophic archaea within these habitats. Anaerobic remineralization within whale-fall environments has been typically described as sulfidogenic (Deming et al., 1997), capable of supporting sulfide-based chemosynthetic communities. Our results, however, suggest that whale-falls are also suitable habitats for diverse methane-producing microbial assemblages, with very little observed production of sulfide. Methanogenic archaea, in particular, are assumed to play only a small role in the breakdown of organic carbon within shallow seafloor sediments compared with aerobic respiration and sulfate reduction, especially at the low temperatures occurring in the deep sea. Our study revealed the presence of a specialized community under the whale-fall, as compared to the surrounding deep-sea floor, dominated by methylotrophic and H2-utilizing methanogens that appear to undergo successional changes over time.
Previous characterization of deep-sea sediments suggest the presence of diverse euryarchaeotal and crenarchaeotal groups (Vetriani et al., 1999). In these studies, up to 60% of clones in typical deep-sea sediments were Crenarchaeota MGI and, similarly, phylotypes representing the Crenarchaeota MBGB have been recovered from subsurface marine sediments (Vetriani et al., 1999; Dhillon et al., 2005; Knittel et al., 2005; Kendall et al., 2007). Similar diversity was recovered in our study, with significant representation by both crenarchaeotal groups (up to 78% of the recovered ribotypes; Figure 5) in ‘reference’ sediments collected ∼10 m from the 2893 m whale-fall, as well as ‘reference’ sediments from a 1000 m whale-fall, which also included the Thermoplasmatales-related marine benthic group D (data not shown). This suggests the widespread occurrence of these archaeal groups in deep-sea sedimentary environments.
In contrast, microbial biomass was enhanced under the whale-fall, with archaea representing 10–40% of the total prokaryote cell counts. Archaea generally only represent 1–12% of the total marine and freshwater microbial diversity; however, in some environments, such as Antarctic sediments, they can comprise up to 34% of the prokaryotic community (Falz et al., 1999; Purdy et al., 2003). Under the whale-fall, these archaea appear to be comprised predominantly of methanogenic members of the Methanomicrobiales (Methanogenium) and Methanosarcinales (Methanosarcina and Methanococcoides), including phylotypes related to those found in other marine environments (Chong et al., 2002; Singh et al., 2005).
Successional changes from methylotrophy to hydrogenotrophy
Insights into potential succession in the methanogenic assemblages directly beneath the whale-fall included the early establishment of methylotrophic methanogens (at 8+ months) followed by an increase in phylotypes related to the hydrogenotrophic Methanogenium (at 33–51+ months). Early in the evolution of the whale-fall, sediments under the whale carcass were dominated by Methanococcoides, an archaeal taxon capable of using C1 compounds, such as methanol and methylamines, and previously recovered from permanently cold marine environments and methane seeps (Singh et al., 2005). Use of these non-competitive substrates presumably enables the early establishment and co-occurrence of methanogenesis and sulfate reduction (Oremland and Polcin, 1982; Winfrey and Ward, 1983). The observation of methylotrophic methanogens from two independent whale-falls may reflect both the production and consumption of methylated non-competitive substrates during the initial stages of degradation. In the latter time points, relatives of the hydrogenotrophic Methanogenium were the dominant methanogen group recovered in samples collected between 33 and 51 months after the discovery of the whale-fall. This particular methanogen comprised the bulk of the archaeal diversity, with minimal representation by members of the Methanosarcinales. Overall, our results support the establishment and persistence of a diversified methanogenic community potentially capable of both competitive (H2/CO2 and acetate) and non-competitive (for example, trimethylamine (TMA)) substrate use. Follow-up studies, including additional analyses of early time points are necessary to determine how much of this pattern is due to natural heterogeneity in sedimentary communities versus true successional development associated with different stages in the evolution of whale-fall habitats (currently in progress).
Pathways of organic carbon remineralization: the role of sulfate
Different groups of prokaryotes dominate the terminal step in carbon catabolism depending on the prevailing environmental conditions (Capone and Kiene, 1988). In the majority of marine environments, greater sulfate concentrations, increased affinity for substrates and more favorable thermodynamic free energy yields results in the dominance of sulfate respiration over methanogenesis. Active sulfate reduction has been demonstrated for sediment sulfate levels as low as 1–3 mM (Boudreau and Westrich, 1984). The lowest sulfate value measured at the 2893 m whale-fall, although depleted compared to the overlying seawater, was 9 mM, a presumably sufficient concentration to support sulfate reduction in the sediments underlying the whale-fall. Preliminary molecular analysis of the bacterial community in these whale-fall sediments (33 months) indicates the occurrence of sulfate reducers, related to members of the Desulfobacterium, Desulfobacula, as well as additional uncultured Deltaproteobacteria (data not shown).
Pathways of organic carbon remineralization: the role of hydrogen
Molecular hydrogen is an important energy source for the anaerobic biodegradation of organic materials (Reeve et al., 1997; Hoehler et al., 2001). Methanogenesis could, in theory, co-occur with sulfate reduction if there was an abundance of competitive substrates, such as hydrogen or acetate. In our study, the presence of H2-utilizing methanogens within the sulfate-replete upper 0–3 cm sediments was surprising in the context of the standard redox gradients in marine sediments. This deviation from standard redox progression may be driven by the surplus of bioavailable carbon. As mentioned previously, the quantity and sources of organic carbon in sediments can also be a major factor determining the distribution of microbes involved in the anaerobic remineralization of carbon. Both lipids and proteins are abundant in a decomposing whale, with specific δ13C isotopic signatures of approximately −24‰ and −15‰, respectively (Stott et al., 1997). Isotopic evidence for preferential utilization of a particular class of whale-derived organic compounds, however, was inconclusive. δ13C values of total carbon were constant, around −20‰, under the whale-fall at all sediment depth horizons and all time points, and thus, difficult to distinguish from typical marine organic matter.
Fermentation results in the production of low molecular weight organic acids, alcohols and molecular hydrogen. Hydrogen enrichment has been shown to stimulate methanogenesis in the presence of sulfate reduction (Winfrey and Zeikus, 1977; Oremland and Taylor, 1978). As the whale carcass degrades, it is reasonable to assume a stimulation in H2 production derived from enhanced fermentation of this concentrated source of organic carbon. We hypothesize that active fermentation provides a surplus of H2 and supports the establishment of hydrogenotrophic methanogens, observed at later time points. Although H2 partial pressures are likely kept low by rapid turnover (Conrad, 1999), we believe H2 production would be continuous due to large-scale fermentation, and potentially in excess of what can be utilized by other microbes, including sulfate reducers. A parallel preliminary study of bacterial communities in these same sediments revealed an abundance of fermentative bacteria, such as Clostridia aceticum and C. purinolyticum, in addition to diverse sulfate-reducing Deltaproteobacteria mentioned above (data not shown). In addition, hydrogen has recently been shown to evolve during methane production via catabolism of C1 compounds, as well (Finke et al., 2007); thus, it is also possible that H2 transfer could occur between the methylotrophic and hydrogenotrophic methanogens observed in this study.
At low temperatures, hydrogenotrophic methane production is thought to be inhibited, shifting methanogenesis toward acetate or C1 compounds (Zeikus and Winfrey, 1976; Gunnarsson and Ronnow, 1982; Schulz and Conrad, 1996; Nozhevnikova et al., 1997). Despite this limitation, a few species of Methanogenium have been observed and even isolated from psychrophilic sulfate-rich marine sediments (Parkes et al., 1990; Franzmann and Liu, 1997; Chong et al., 2002; Purdy et al., 2003; Kendall and Boone, 2006). High abundance of the methyl-coenzyme M reductase (mcrA) gene was documented in Methanogenium-dominated sediments in this study, suggesting that despite low temperatures, methanogenic activity belonging to H2-utilizing methanogens may occur in sediments underlying the whale-fall.
Evolution of the methane cycle from production to consumption
The diverse methanogenic archaea detected under the whale-fall are presumably responsible for the elevated methane concentrations (up to 4 mmol g−1 sediments CH4) measured at these organically enriched near-seafloor habitats. Indirect evidence of active methane production affiliated with deep-sea whale-falls has been reported in a previous study of a whale-fall at ∼4000 m depth near the Torishima Seamount, Japan (Naganuma et al., 1996). The presence of methane-oxidizing bacteria, based on the analysis of fatty-acid methyl esters, was suggested; however, neither the occurrence of methanogens nor methane concentrations were reported (Naganuma et al., 1996). In our study, we not only detected methanogenic populations but we also observed geochemical and phylogenetic evidence of the anaerobic oxidation of methane. In a single sediment core taken at 1 m distance from the whale-fall (PC47), phylotypes related to ANME-3 archaea (determined by both 16S rRNA and mcrA analysis) dominated the 9–12 cm depth horizon, while members of the Methanococcoides were abundant in the 3–6 cm depth horizon, suggesting active methanogenesis overlying a deeper horizon of methane oxidation. Accordingly, the lightest δ13CH4 value (−58‰) corresponded to the depth horizon dominated by Methanococcoides (Tables 1 and 2 and Figure 2a). The depleted δ13CH4 value in the Methanococcoides-dominated depth layer is consistent with light methane produced by pure cultures of Methanococcoides burtonii during transformation of trimethylamine to methane (Summons et al., 1998). The heaviest value (−39‰), on the other hand, corresponded to the horizon dominated by the ANME-3 group (Tables 1 and 2 and Figure 2a). The observed 19‰ shift in the methane isotopic signature is likely due to preferential microbial consumption of the lighter molecule by the ANME-3 group during the anaerobic oxidation of methane. Fractionation factors for methane in the presence of methane-oxidizing ANME-3 archaea have been recently observed to be in this range (T Holler, Max Planck Institute, personal communication), although other factors affecting biological fractionation such as substrate availability may also play a role. Additional evidence for active methanotrophy at depth was expressed in the δ13C-TOC from the ANME-3-dominated sediment horizon, with a ∼2‰ depletion in TOC (−22.2‰ versus −18.7‰ for the deeper and shallow sediment horizons, respectively; Figure 2b). The depth distribution of methanogenic and methanotrophic archaea within the whale-fall habitat is in contrast to what has been previously reported from marine methane seeps, where the source for methane typically originates from deeper sediment horizons and methanotrophic archaea typically colonize the overlying sulfate-replete sediments (Orphan et al., 2001; Knittel et al., 2005). The inverted distribution in methanogenic and methanotrophic archaea observed in our study is likely a result of the substantial organic enrichment created in the surface sediments within and adjacent to the whale-fall.
Translocation of organic carbon: radius of influence
At the 2893 m whale-fall, the enrichment and lateral extent of whale-derived total sediment carbon appeared to reach the surrounding sea floor to at least 10 m, forming a ‘bulls eye’ around the whale-fall. By 33 months from the time of discovery, the whale-fall was estimated to have already been on the sea floor for 6–12 months, thus the rate and full extent of Corg translocation were not known. Interestingly, lateral organic enrichment appeared to be far greater than enrichment with increasing sediment depth immediately under the whale-fall. This transport of carbon from the site of origin, apparently confined to the upper 3 cm, is likely due to bioturbation by megafauna in the area (Braby et al., 2007). For the 2893 m whale-fall, no noticeable change in TOC was observed after 8 months post-discovery. Whatever the specific time frame for TOC enrichment, Corg (and even Norg) under and immediately surrounding the 2893 m whale-fall are much higher than typical deep-sea sediments. Values for sedimentary organic carbon and nitrogen from Monterey Canyon have been reported, averaging 0.5% for TOC and 0.05% for total organic nitrogen (Paull et al., 2006). These measurements, made at depths of only 600 m, are likely over estimates of the conditions occurring at 2893 m, suggesting that the whale-fall has left a large footprint with elevated, but heterogeneous, organic matter associated with this unique seafloor habitat. A more detailed investigation of the timing and spatial extent of organic enrichment in relation to the whale-fall should be conducted.
Conclusion
Both 16S rRNA and mcrA surveys indicate the presence of phylotypes related to the metabolically versatile C1-utilizing Methanococcoides as well as the H2-utilizing Methanogenium in sediments underlying a deep-sea whale-fall in Monterey Canyon. Temporal changes in community structure included the early establishment of methylotrophic methanogens followed by later development of methanogens thought to be hydrogenotrophic. Our results suggest elevated concentrations of bioavailable carbon, creating overlapping niches for methanogenic archaea and sulfate-reducing bacteria and, thus, a potential role for methanogens in the overall oxidation of enriched sources of organic matter in permanently cold marine sediments. The existence of methanogens within sulfate-rich sediments underlying whale carcasses suggests atypical anaerobic remineralization pathways, possibly supported by enhanced rates of fermentation and associated H2 production. It is currently unknown how much of the organic carbon respiration at whale-falls is accounted for by methanogenesis but it is clear that they are an important point source of methane generation in shallow seafloor sediments, with levels of methanogenic activity sufficient enough to support the subsequent establishment of methane-oxidizing archaea and a complete and active near-seafloor methane cycle within these environments. The assessment of electron flow and the turnover of competitive versus non-competitive substrates was not the focus of this investigation; however, contextual measurement of these compounds, specifically hydrogen, C1 and C2 compounds, is a necessary next step in understanding terminal carbon metabolism in this environment. Future work building on these initial observations will incorporate microbial rate measurements and additional biogeochemical analyses to assist in further constraining the local physicochemical environment supporting enhanced microbial activity and methane production in this unusual seafloor environment.
References
Association APH (1976). Standard Methods for the Examination of Water and Waste Water, 14th edn. American Public Health Association, Inc.: Washington, DC.
Barry JP, Greene HG, Orange DL, Baxter CH, Robison BH, Kochevar RE et al. (1996). Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep Sea Res I 43: 1739–1762.
Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM . (1997). Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA 94: 277–282.
Boudreau BP, Westrich JT . (1984). The dependence of bacterial sulfate reduction on sulfate concentration in marine sediments. Geochim Cosmochim Acta 48: 2503–2516.
Braby CE, Rouse GW, Johnson SB, Jones WJ, Vrijenhoek RC . (2007). Bathymetric and temporal variation among Osedax boneworms and associated megafauna on whale-falls in Monterey Bay, California. Deep Sea Res I 54: 1773–1791.
Capone DG, Kiene RP . (1988). Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon catabolism. Limnol Oceanogr 33: 725–749.
Capone DG, Reese DD, Kiene RP . (1983). Effects of metals on methanogenesis, sulfate reduction, carbon dioxide evolution, and microbial biomass in anoxic salt marsh sediments. Appl Environ Microbiol 45: 1586–1591.
Cappenberg TE . (1974). Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a fresh-water lake. I. Field observations. Antonie Van Leeuwenhoek 40: 285–295.
Chong SC, Liu Y, Cummins M, Valentine DL, Boone DR . (2002). Methanogenium marinum sp. nov., a H2-using methanogen from Skan Bay, Alaska, and kinetics of H2 utilization. Antonie van Leeuwenhoek 81: 263–270.
Cline JD . (1969). Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14: 454–458.
Conrad R . (1999). Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol Ecol 28: 193–202.
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M . (1999). The domain-specific probe Eub338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444.
Delong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
Deming JW, Reysenbach A-L, Macko SA, Smith CR . (1997). Evidence for the microbial basis of a chemoautotrophic invertebrate community at a whale-fall on the deep seafloor: bone-colonizing bacteria and invertebrate endosymbionts. Microsc Res Tech 37: 162–170.
Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A . (2005). Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol 71: 4592–4601.
Falz KZ, Holliger C, Grosskopf R, Liesack W, Nozhevnikova AN, Muller B et al. (1999). Vertical distribution of methanogens in the anoxic sediment of Rotsee (Switzerland). Appl Environ Microbiol 65: 2402–2408.
Finke N, Hoehler TM, Jorgensen BB . (2007). Hydrogen ‘leakage’ during methanogenesis from methanol and methylamine: implications for anaerobic carbon degradation pathways in aquatic sediments. Environ Microbiol 9: 1060–1071.
Franzmann PD, Liu Y . (1997). Methanogenium frigidum sp. nov., a psychrophilic, H2-using methanogen from Ace Lake, Antarctica. Int J Syst Bacteriol 47: 1068–1072.
Gieskes JM, Gamo T, Brumsack H . (1991). Chemical methods for interstitial water analysis aboard JOIDES Resolution. Ocean Drilling Program Texas A&M University: Texas, Technical Note, vol. 15. pp 24–33.
Girguis PR, Orphan VJ, Hallam SJ, Delong EF . (2003). Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl Environ Microbiol 69: 5472–5482.
Goffredi SK, Paull CK, Fulton-Bennett K, Hurtado LA, Vrijenhoek RC . (2004). Unusual benthic fauna associated with a whale-fall in Monterey Canyon, California. Deep Sea Res I 51: 1295–1306.
Gunnarsson LAH, Ronnow PH . (1982). Interrelationships between sulfate reducing and methane producing bacteria in coastal sediments with intense sulfide production. Mar Biol 69: 121–128.
Hoehler TM, Alperin MJ, Albert DB, Martens CS . (2001). Apparent minimum free energy requirements for methanogenic archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol Ecol 38: 33–41.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103: 2815–2820.
Jørgensen BB . (1982). Mineralization of organic matter in the sea bed––the role of sulphate reduction. Nature 296: 643–645.
Jørgensen BB, Boetius A . (2007). Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol 5: 770–781.
Kendall MM, Boone DR . (2006). Cultivation of methanogens from shallow marine sediments at Hydrate Ridge, Oregon. Archaea 2: 31–38.
Kendall MM, Wardlaw GD, Tang CF, Bonin AS, Liu Y, Valentine DL . (2007). Diversity of archaea in marine sediments from Skan Bay, Alaska, including cultivated methanogens, and description of Methanogenium boonei sp. nov. Appl Environ Microbiol 73: 407–414.
Kennett J . (1982). Marine Geology. Prentice-Hall Inc.: Englewood Cliffs, NJ.
Knittel K, Losekann T, Boetius A, Kort R, Amann R . (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71: 467–479.
Kristjansson JK, Schönheit P, Thauer RK . (1982). Different Ks values for hydrogen of methanogenic bacteria and sulfate reducing bacteria: an explanation for the apparent inhibition of methanogenesis by sulfate. Arch Microbiol 131: 278–282.
Li L, Kato C, Horikoshi K . (1999). Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol 1: 391–400.
Losekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl Environ Microbiol 73: 3348–3362.
Lovley DR, Dwyer DF, Klug MJ . (1982). Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Appl Environ Microbiol 43: 1373–1379.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar AB et al. (2004). ARB: a software environment for sequence data. Nucleic Acid Res 32: 1363–1371.
Luton PE, Wayne JM, Sharp RJ, Riley PW . (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148: 3521–3530.
Massana R, DeLong EF, Pedros-Alio C . (2000). A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66: 1777–1787.
Mountfort DO, Asher RA . (1979). Effect of inorganic sulfide on the growth and metabolism of Methanosarcina barkeri strain DM. Appl Environ Microbiol 37: 670–675.
Naganuma T, Wada H, Fujioka K . (1996). Biological community and sediment fatty acids associated with the deep-sea whale skeleton at the Torishima Seamount. J Oceanogr 52: 1–15.
Niemann H, Losekann T, de Beer D, Elvert M, Nadalig T, Knittel K et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443: 854–858.
Nozhevnikova AN, Holliger C, Ammann A, Zehnder AJB . (1997). Methanogenesis in sediments from deep lakes at different temperatures (2–70°C). Water Sci Technol 36: 57–64.
Oremland RS, Marsh LM, Polcin S . (1982). Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments. Nature 296: 143–145.
Oremland RS, Polcin S . (1982). Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol 44: 1270–1276.
Oremland RS, Taylor BF . (1978). Sulfate reduction and methanogenesis in marine sediments. Geochim Cosmochim Acta 42: 209–214.
Orphan VJ, Hinrichs KU, Ussler WI, Paull CK, Taylor LT, Sylva SP et al. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediment. Appl Environ Microbiol 67: 1922–1934.
Parkes RJ, Cragg BA, Fry JC, Herbert RA, Wimpenny JWT . (1990). Bacterial biomass and activity in deep sediment layers from the Peru margin. Philos Trans R Soc London B Biol Sci 331: 139–153.
Paull CK, Ussler III W, Mitts PJ, Caress DW, West GJ . (2006). Discordant 14C-stratigraphies in upper Monterey Canyon: a signal of anthropogenic disturbance. Mar Geol 233: 21–36.
Pernthaler J, Amann R, Pernthaler A . (2004). Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. In: Kowalchuk G, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD (eds). Molecular Microbial Ecology Manual. Kluwer Academic Publishers: London, pp 711–725.
Purdy KJ, Nedwell DB, Embley TM . (2003). Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl Environ Microbiol 69: 3181–3191.
Reeburgh WS, Heggie DT . (1977). Microbial methane consumption reactions and their effects on methane distributions on freshwater and marine environments. Limnol Oceanogr 22: 1–9.
Reeve JN, Morgan RN, Nolling J . (1997). Environmental and molecular regulation of methanogenesis. Water Sci Technol 36: 1–6.
Schulz S, Conrad R . (1996). Influence of temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microbiol Ecol 20: 1–14.
Senior E, Lindstrom EB, Banat IM, Nedwell DB . (1982). Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the east coast of the United Kingdom. Appl Environ Microbiol 43: 987–996.
Shepard FP . (1973). Submarine Geology. Harper & Row: New York, 517pp.
Singh N, Kendall MM, Liu Y, Boone DR . (2005). Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica. Int J Syst Evol Microbiol 55: 2531–2538.
Smith CR, Baco AR . (2003). The ecology of whale-falls at the deep-sea floor. Oceanogr Mar Biol Annu Rev 41: 311–354.
Stahl DA, Amann R . (1991). Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons Ltd: Chichester, England, pp 205–248.
Stein LY, Jones G, Alexander B, Elmund K, Wright-Jones C, Nealson KH . (2002). Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol Ecol 42: 431–440.
Stott AW, Evershed RP, Tuross N . (1997). Compound specific approach to the δ13C analysis of cholesterol in fossil bones. Org Geochem 26: 99–103.
Summons RE, Franzmann PD, Nichols PD . (1998). Carbon isotopic fractionation associated with methylotrophic methanogenesis. Org Geochem 28: 465–475.
Swofford DL . (1998). PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Sinauer: Sunderland, MA.
van der Maarel MJ, Artz RR, Haanstra R, Forney LJ . (1998). Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol 64: 2894–2898.
Van Dover CL . (2000). The Ecology of Deep-Sea Hydrothermal Vents. Princeton University Press: Princeton, 411pp.
Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach A-L . (1999). Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol 65: 4375–4384.
Visscher PT, Baumgartner LK, Buckley DH, Rogers DR, Hogan ME, Raleigh CD et al. (2003). Dimethyl sulphide and methane thiol formation in microbial mats: potential pathways for biogenic signatures. Environ Microbiol 5: 296–308.
Winfrey MR, Ward DM . (1983). Substrates for sulfate reduction and methane production in intertidal sediments. Appl Environ Microbiol 45: 193–199.
Winfrey MR, Zeikus JG . (1977). Effect of sulfate on carbon and electron flow during microbial methanogenesis in freshwater sediments. Appl Environ Microbiol 33: 275–281.
Zeikus JG, Winfrey MR . (1976). Temperature limitation of methanogenesis in aquatic sediments. Appl Environ Microbiol 31: 99–107.
Acknowledgements
This study was supported by a Davidow grant to Caltech's Geological and Planetary Sciences division, US National Science Foundation (MCB-0454860 to SKG), and the Gordon and Betty Moore Foundation (to VJO). We thank the Tiburon pilots and Western Flyer crew and chief scientist R Vrijenhoek for allowing our participation in research cruises; S Johnson and WJ Jones for shipboard support; A Pernthaler for advice regarding FISH analyses; B Ussler for help with methane measurements; Patty Tavormina for QPCR data; Tsegereda Embaye for CARD FISH preparations and B Harrison for ARB assistance.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Goffredi, S., Wilpiszeski, R., Lee, R. et al. Temporal evolution of methane cycling and phylogenetic diversity of archaea in sediments from a deep-sea whale-fall in Monterey Canyon, California. ISME J 2, 204–220 (2008). https://doi.org/10.1038/ismej.2007.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.103
Keywords
This article is cited by
-
In-situ observations of an intact natural whale fall in Palmer deep, Western Antarctic Peninsula
Polar Biology (2023)
-
Phylogenetic characterization of marine microbial biofilms associated with mammal bones in temperate and polar areas
Marine Biodiversity (2020)
-
Microbial insights from Antarctic and Mediterranean shallow-water bone-eating worms
Polar Biology (2020)
-
In situ development of a methanotrophic microbiome in deep-sea sediments
The ISME Journal (2019)
-
Variation of geochemical environments associated with whale-fall biomass mineralization processes in the sediment during the mobile scavenger, enrichment opportunist, and sulfophilic stages
Marine Biology (2018)