Abstract
In the present study, we investigate the expression profile of the epidermal growth factor receptor family, which comprises EGFR/ErbB1, HER2/ErbB2, HER3/ErbB3 and HER4/ErbB4 in oral leukoplakia (LP). The expression of four epidermal growth factor receptor (EGFR) family genes and their ligands were measured in LP tissues from 14 patients and compared with levels in 10 patients with oral lichen planus (OLP) and normal oral mucosa (NOM) from 14 healthy donors by real-time polymerase chain reaction (PCR) and immunohistochemistry. Synchronous mRNA coexpression of ErbB1, ErbB2, ErbB3 and ErbB4 was detected in LP lesions. Out of the receptors, only ErbB4 mRNA and protein was more highly expressed in LP compared with NOM tissues. These were strongly expressed by epithelial keratinocytes in LP lesions, as shown by immunohistochemistry. Regarding the ligands, the mRNA of Neuregulin2 and 4 were more highly expressed in OLP compared with NOM tissues. Therefore, enhanced ErbB4 on the keratinocytes and synchronous modulation of EGFR family genes may contribute to the pathogenesis and carcinogenesis of LP.
Similar content being viewed by others
Introduction
Leukoplakia (LP) is defined as a white patch or plaque which cannot be characterized clinically or pathologically as any other disease. Because LP has the potential for malignant alteration, it is important for clinicians to observe changes in clinical features and routine histological investigations in LP lesions. It has been suggested that it may undergo malignant transformation as the severity in epithelial dysplasia increases,1,2 while other investigators have suggested that the degree of dysplasia in LP is not related to the propensity to undergo malignant transformation.3 As yet, there has been a failure to identify markers, either in isolation or as a panel, which allows for a prognostic prediction of oral pre-cancer which is any more reliable than dysplasia recording. Therefore, there is a substantial need to improve the histological assessment of epithelial dysplasia. Alternatively, it may be necessary to find genomic markers for predicting the malignant potential of LP lesions.
The proto-oncogene referred to as epidermal growth factor receptor (EGFR) is a well-known tyrosine kinase growth factor receptor. The EGFR family comprises four distinct receptors: EGFR/ErbB1, HER2/ErbB2, HER3/ErbB3 and HER4/ErbB4. Binding of a ligand to the receptor and the subsequent receptor hetero and homo dimerization leads to a phosphorylation cascade mediated via tyrosine kinases.4 Abnormal amplification of the ErbB1 gene has been observed widely in various human tumors, including lung carcinoma, laryngeal and oral squamous cell carcinoma (SCC).5,6,7 It has been shown that the colocalization of multiple EGFR family were detected in oral SCC.8 The expression of ErbB1 is localized to the basal layer in normal oral mucosa (NOM), while it is widely distributed in the spinous layer in LP.9 However, a detailed evaluation describing the expression of the EGFR family in LP tissues has not been reported so far.
In the present study, to clarify the expression profile of EGFR family genes in LP, we determined the expression of four members of the EGFR family (ErbB1, ErbB2, ErbB3 and ErbB4), 7 major EGF-like growth factors (epidermal growth factor, EGF; amphiregulin, AREG; transforming growth factor-α, TGF-α; epiregulin, EREG; heparin-binding EGF-like growth factor, HB-EGF; Neuregulin, NRG1–4; and betacellulin, BTC) in the oral epithelium of biopsy specimens of patients with LP using quantitative real-time polymerase chain reaction (PCR) and immunohistochemistry.
Materials and methods
Patients and samples
LP tissues were obtained from 14 LP patients (seven men, seven women, mean age: 62.5±11.1 years) (Table 1). Demographic data, personal habits of tobacco smoking and alcohol consumption and clinical features of lesions were also recorded. LP patients had no history of metal allergies, local or systemic allergic drug reactions, and had not received topical or systemic steroid therapy. All of the LP samples were clinically defined as the homogeneous type, and pathological diagnosis, including the degree of dysplasia (without, mild, moderate and severe), was confirmed by the same pathologist. Erythroplakia, speckled LP, malignant transformation of LP or early carcinomas in situ were excluded from this study. As comparative lesions, oral lichen planus (OLP) tissues were obtained from 10 OLP patients (five men, five women, mean age: 55.8±15.3 years) (Table 2). OLP patients were included in this study if they presented with a histological diagnosis of OLP, had no history of metal allergies, or local or systemic allergic drug reactions, and had not received topical or systemic steroid therapy. All of the OLP samples were the reticular type. The LP and OLP tissue specimens were taken before been divided into two samples. One was used for the histopathological or immunohistochemical study, and the other was used for RNA extraction. As control samples, the biopsy specimens obtained from NOM of 14 healthy donors (seven men, seven women, mean age: 26.5±10.6 years) was taken in the same manner. The healthy donors had not experienced any diseases of the oral mucosa.
The study was approved by the Research Ethical Committee of Tsurumi University, Yokohama Rosai Hospital and National Hospital Organization, Sagamihara National Hospital, and informed consent for this study was obtained from all of the patients and healthy donors.
Preparation of tissue specimens
After biopsy, one half of each tissue specimen was immediately soaked in RNAlater RNA Stabilization Reagent (Qiagen, Tokyo, Japan) and stored at −20 °C until RNA extraction.
RNA extraction and cDNA synthesis
NOM, OLP and LP tissues were homogenized and RNA was extracted using the RNeasy lipid tissue kit (Qiagen). Total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit (Takara Bio, Shiga, Japan). One-tenth of the cDNA was amplified with the SYBR Premix Ex Taq RT-PCR Kit (Takara Bio) according to the manufacturer’s instructions.
Quantitative RT-PCR
Real-time PCR was performed using a LightCycler 2.0 (Roche Diagnostics, Tokyo, Japan) following the manufacturer’s protocol. The PCR products were assessed by the fluorescence from the SYBR Green that intercalated to the DNA fragments, and melting curves were routinely recorded to verify the singularity of the products. The primer sequences of the genes tested in this study were: F 5′-GGTGCGAATGACAGTAGCATTATGA-3′, R 5′-AAAGGTGGGCTCCTAACTAGCTGAA-3′ (ErbB1); F 5′-CAGGCACCGCAGCTCATCTA-3′, R 5′-TCCCAGGTCACCATCAAATACATC-3′ (ErbB2); F 5′-CCCAGCATCTGAGCAAGGGTA-3′, R 5′-TTTAGGCGGGCATAATGGACA-3′ (ErbB3); and F 5′-TGATAGGCCGTTGGTTGTCTGA-3′, R 5′-CCAGGTAGACATACCCAATCCAGTG-3′ (ErbB4). F 5′-GTGATTCCATCATGTATCCCAGGAG-3′, R 5′-AGATGCACTGTCCATGCAAACAA-3′ (EREG); F 5′-CTTCACTGTGTGGTGGCAGATG-3′, R 5′-ATGCAGTAATGCTTGTATTGCTTGG-3′ (BTC); F 5′-CAACCAGTGGCTGGTGAGGA-3′, R 5′-GAGCCCTTATCACTGGATACTGGAA-3′ (EGF); F 5′-GTGGTGCTGTCGCTCTTGATACTC-3′, R 5′-TCAAATCCATCAGCACTGTGGTC-3′ (AREG); F 5′-AGATAGACAGCAGCCAACCCTGA-3′, R 5′-CTAGGGCCATTCTGCCCATC-3′ (TGF-a); F 5′-GGGCATGACTAATTCCCACTGA-3′, R 5′-GCCCAATCCTAGACGGCAAC-3′ (HBEGF); F 5′-TCGGTGTGAAACCAGTTCTGAATA-3′, R 5′-TCTCCAGAATCAGCCAGTGATG-3′ (NRG1); F 5′-ACCCTAGGCTTGGAGCTGGA-3′, R 5′-CCATTCGGGTAGCTGTGTCTTTATC-3′ (NRG2); F 5′-GTGGATGCACCAGTGAGCAG-3′, R 5′-CTGTGATGAGGCTATTGGGTGAA-3′ (NRG3); F 5′-ATCAGTCCATCGGCAACTGCTA-3′, R 5′-ACATTTGCCTCTGGTTGCTTCA-3′ (NRG4). The amplified product using each primer pair was cloned into the pGEM-T vector (Promega, Tokyo, Japan) and the plasmid was linearized by enzymatic digestion for use as the quantitation standard. A reference cDNA was used in every assay to assess the interassay precision. The cDNA levels among the samples were normalized to the expression level of the internal control gene, GAPDH, (F 5′-GCACCGTCAAGGCTGAGAAC-3′, R 5′-ATGGTGGTGAAGACGCCAGT-3′).
The expressions of the EGFR family genes in the NOM, OLP and LP tissues and the expressions of EGFR family ligands genes in LP were analyzed by the Mann–Whitney U test, and the correlations between EGFR family members were analyzed by Spearman’s rank correlation coefficient. Moreover, the relationships between the expressions of the EGFR family genes and the degree of dysplasia in LP were examined by Spearman’s rank correlation coefficient.
Histology and immunohistochemistry
For histopathology, 4-μm sections of paraffin-embedded NOM, OLP and LP tissue specimens were stained with hematoxylin and eosin (HE). Using commercially available monoclonal or polyclonal antibodies and following the ENVISION technique (K4003; Dako, Copenhagen, Denmark) with 3,3′-diaminobenzidine tetrahydrochloride as the indicator agent, immunohistochemical analyses were performed for the expressions of the following EGFR family: ErbB1 (k1492; Dako, Copenhagen, Denmark), ErbB2 (427041; Nichirei, Tokyo, Japan), ErbB3 (ab15136; Abcam PLC, Cambridge, UK) and ErbB4 (sc-283; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Statistical analysis
The statistical analysis was carried out using StatView statistical analysis software (SAS, Cary, CA, USA). Differences between the NOM, OLP and LP tissues were determined to be significant when P<0.05 using a Mann–Whitney U test and Spearman’s rank correlation coefficient.
Results
Gene expression profiles of EGFR family and its ligands
We determined the mRNA expression level of EGFR family in NOM, OLP and LP tissues by real-time PCR. The expression levels of GAPDH were quite equal in all tissue specimens, and therefore, the cDNA levels among the samples were normalized by the corresponding expression levels of GAPDH as an internal control gene. Out of the four EGFR family, ErbB1-4, the expression of ErbB4 mRNA was significantly higher in LP compared with NOM (P<0.05), and the expression of ErbB1 mRNA was significantly higher in OLP compared with NOM (P<0.05) (Figure 1a). There were no significant differences in ErbB1, ErbB2, ErbB3 and ErbB4 expression between LP and OLP. Moreover, we also determined the mRNA expression level of EGFR family ligands in LP tissues by real-time PCR. The expression of NRG2 and NRG4 mRNA were significantly higher in LP compared with NOM (P<0.05), and the expression of BTC mRNA was significantly higher in NOM compared with LP (P<0.05) (Figure 1b).
Gene expression profiles of EGFR family and its ligands. The expression mRNA levels of EGFR family and its ligands were measured by real-time quantitative PCR. Each dot indicates a single sample. The GAPDH gene was used as an internal control. (a) EGFR family genes. Out of four EGFR family, the expression of ErbB1 mRNA was significantly higher in OLP compared with NOM and the expression of ErbB4 mRNA was significantly higher in LP compared with NOM (P<0.05). (b) EGFR family ligands genes. The expression of NRG2 and NRG4 mRNA was significantly higher in LP compared with NOM (P<0.05), and the expression of BTC mRNA was significantly higher in NOM compared with LP (P<0.05). *P<0.05, **P<0.01. BTC, betacellulin; EGFR, epidermal growth factor receptor; LP, leukoplakia; NOM, normal oral mucosa; OLP, oral lichen planus; PCR, polymerase chain reaction.
Protein expression profiles of EGFR family in NOM, OLP and LP
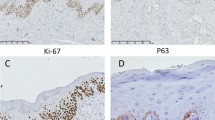
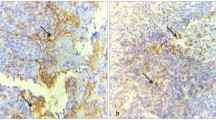
hematoxylin and eosin staining of OLP tissue specimens showed lichenoid degeneration of the basal layer of the epithelium with band-like infiltrates of lymphocytes in the superficial lamina propria (Figure 2B and 2b). The LP tissue specimens revealed a keratinized layer of surface epithelium (hyperkeratosis), a thickened spinous layer (acanthosis) and basilar cells. (Figure 2C and 2c). We determined the protein expression of EGFR family members in NOM, OLP and LP tissues by immunohistochemistry. Expression of ErbB1 was detected in LP, OLP and NOM tissue specimens (Figure 2d–2f), while those of ErbB2, ErbB3 were not seen in OLP, but were slightly expressed in NOM and LP (Figure 2g–2l). In NOM, ErbB1positive cells were mainly keratinocytes of the basal and suprabasal layer (Figure 2d). In LP, the ErbB4 positive cells were mainly in the basal layer and in the spinous layer (Figure 2o).
Protein expression profiles of EGFR family in NOM, OLP and LP. Routine staining for HE and immunohistochemical analysis for the expression of ErbB1, ErbB2, ErbB3 and ErbB4 in NOM, OLP and LP tissues using commercially available monoclonal antibodies and the Envision technique using DAB as an indicator agent. Expression of ErbB1 and ErbB4 was detected in all tissue specimens, while those of ErbB2 and ErbB3 were slightly expressed in NOM, and LP. Magnifications: ×40 (A, B and C) and ×100 (a-o).* indicates basal cell layer; ⋆ indicates prickle cell layer; ★ indicates keratinized layer. DAB, 3,3′-diaminobenzidine tetrahydrochloride; EGFR, epidermal growth factor receptor; HE, hematoxylin and eosin; LP, leukoplakia; NOM, normal oral mucosa; OLP, oral lichen planus.
Correlation between the expression of EGFR family genes and the degree of dysplasia in LP
The expression levels of ErbB4 in LP lesions were significantly correlated with the dysplasia grades (P<0.05), while ErbB1 showed a significant inverse correlation (P<0.05) (Figure 3). No significant correlations between the expression levels of ErbB2 or ErbB3 and dysplasia grades were detected.
Correlation between the expression of EGFR family genes and the degree of dysplasia in LP. All of the LP samples were clinically defined as the homogeneous type, and pathological diagnosis including the degree of dysplasia was confirmed by the same pathologist. The GAPDH gene was used as an internal control. The expression levels of ErbB4 were significantly correlated with the dysplasia grades (P<0.05), while ErbB1 showed a significant inverse correlation (P<0.05). No significant correlations between the expression levels of ErbB2 or ErbB3 and dysplasia grades were detected. Spearman’s rank correlation coefficient was used, with P<0.05 regarded as statistically significant. EGFR, epidermal growth factor receptor; LP, leukoplakia.
Precedence and coexpression of EGFR family genes in NOM, OLP and LP
ErbB2 mRNA was the most highly expressed of the EGFR family members, followed by ErbB3, ErbB1, and ErbB4 in NOM, OLP and LP tissue specimens (Figure 4). There are no significant differences in EGFR family gene expression between NOM, OLP and LP tissues. With regard to the coexpression of EGFR family genes, the mRNA expression levels of ErbB2 and ErbB3 were significantly correlated in NOM, OLP and LP tissues (Figure 5). Furthermore, there was also a significant correlation between the mRNA expression levels of ErbB1 with ErbB2 (r=0.890, P<0.05) and ErbB3 (r=0.727, P<0.05) in LP tissues (Figure 5), but not with ErbB4 (data not shown).
Precedence of EGFR family members’ gene expression levels in NOM, OLP and LP. Each point indicates the ErbB1–4 mRNA expression levels. ErbB2 mRNA expression levels were the highest, followed by ErbB3, ErbB1 and ErbB4 in NOM, OLP and LP. There are no significant differences in the precedence gene expression of EGFR family members between NOM, OLP and LP tissues. EGFR, epidermal growth factor receptor; LP, leukoplakia; NOM, normal oral mucosa; OLP, oral lichen planus.
Correlation of mRNA expression levels between ErbB1–4 in NOM, OLP and LP. The GAPDH gene was used as an internal control. The expression levels of ErbB2 and that of ErbB3 significantly correlated in NOM, OLP and LP. Furthermore, there was a significant correlation between the expression levels of ErbB1 with ErbB2 (r=0.890, P<0.05) and ErbB3 (r=0.727, P<0.05) in LP. Spearman’s rank correlation coefficient was used, with P<0.05 regarded as statistically significant. LP, leukoplakia; NOM, normal oral mucosa; OLP, oral lichen planus.
Discussion
In the present study, we showed which of the EGFR family genes were coexpressed, and also demonstrated that ErbB4 expression was increased in the keratinocytes in LP lesions compared with NOM tissues. This suggests that keratinocytes in LP may be more sensitive to stimulation with the ligands for ErbB4, and synchronous modulation of EGFR family genes may be important in the pathogenesis of LP.
The expression of ErbB4 mRNA was significantly higher in LP compared with NOM, and the ErbB4-positive cells were detected in the basal layer and spinous layer in LP. ErbB4 is activated by members of the neuregulin family, and by some members of the EGF growth factor family; BTC, EREG, HB-EGF and NRG family.10 Interestingly, we also demonstrated that the expressions of NRG2 and NRG4 mRNA were significantly higher in LP compared with NOM. Binding of a ligand to the receptor and the subsequent receptor hetero and homo dimerization leads to a phosphorylation cascade mediated via tyrosine kinases;5 therefore, the precise mechanisms regarding the phosphorylation cascade in LP require further investigation.
The role of ErbB4 is less clear, but recent studies have suggested that its ability to form heterodimers with other EGFR family members seems to enhance proliferation and invasion of oral SCC cells.11,12,13,14,15,16 A previous study has also indicated that the expression of all four receptors is associated with shortened survival in patients with oral SCC, with the combination of ErbB1, ErbB2 and ErbB3 (but not ErbB4) giving the greatest prognostic information.8 Moreover, high levels of expression of NRG2 and NRG4 have been reported in human high-grade breast tumors.17 The ErbB4 amplification by the regulation of NRG-mediated signaling may cause changes in the activation of other cell surface proto-oncogene proteins and/or the transcription of other proto-oncogenes, thereby distorting the control of epithelial cell growth and eventually leading to a malignancy.
In ErbB1 expression, abnormal amplification of the ErbB1 gene has been observed widely in various human tumors.5,6,7 It has been shown that the expression of ErbB1 is localized to the basal layer in NOM and is widely distributed in the spinous layer in LP.9 We have previously reported that the enhanced ErbB1 expression on the keratinocytes in OLP lesions and the upregulation of EGF-like ligands in keratinocytes and infiltrating mononuclear cells could contribute to the carcinogenesis and pathogenesis of OLP.18 In this study, there was no significantly higher expression levels of ErbB1 in LP compared with NOM. However, the expression levels of ErbB1 showed a significant inverse correlation with the degree of dysplasia. In the normal state, ErbB1 is expressed in the basal layer where cells proliferate constantly, while it is expressed in the upper spinous layer in abnormal conditions, such as OLP or LP. Histopathology of LP shows the thickened surface layer of parakeratin. ErbB1 is essential for keratinocyte proliferation; therefore, it is possible that upregulated ErbB1 expression is involved in the mechanisms of keratinocyte proliferation. Taking these results together, we suggest that synchronous modulation of the EGFR family genes and ErbB4 may play a key role in the pathogenesis of LP.
Otherwise, previous study has shown a discrepancy between the mRNA levels and protein levels of EGFR family members in oral dysplastic and malignant oral epithelial tissues.8,12,19,20,21 In our study, although the ErbB2 protein expressions in LP were slightly detected in basal layer and pickle layer cells, ErbB2 mRNA was most highly expressed. All of the samples were totally homogenized for RNA extraction; therefore, it is possible that this discrepancy may be due to the extraction of RNA from tissue specimens containing subepithelial connective tissue. The application of laser microdissection technique would represent a valid gene expression profiles in LP; however, the precise evaluation of EGFR family in LP requires further investigation.
In view of the degree of dysplasia in LP, the expression levels of ErbB4 showed a significant correlation. However, ErbB1 was downregulated as the degree of dysplasia increased. A set of criteria for diagnosing epithelial dysplasia has been known for more than 30 years, and it has been repeatedly shown that the evaluation and grading of epithelial dysplasia is very subjective, with poor to moderate agreement rates.2 The histopathological features of dysplasia were individual cell keratinization, bizarre mitoses and mitoses at an abnormally superficial level in the epithelium, the first two of which were among features most important in distinguishing LP undergoing malignant development from other leukoplakias.22 Recent studies have shown that activated ErbB4 dimers support cell growth and mitosis through activation of the p21Ras/Raf-1/MEK/EPK pathway.23 Combinations of several molecular/genetic markers and histopathological features have allowed better understanding of the events characterizing the different stages of cancer development.
It is well established that SCC develops from LP lesions, yet the mechanism of carcinogenesis in LP is still unclear. The concept of a two-step process of cancer development in the oral mucosa, which is the initial presence of a precursor (pre-malignant, pre-cancerous) lesion subsequently developing into cancer, is not well established. Further studies based on the analysis of signaling pathways will promote our understanding of the mechanisms of LP and it may be important to evaluate the combined significance of several markers and/or clinical and histological variables for their prognostic value.
Conclusion
Enhanced expression of ErbB4 on keratinocytes could contribute to the pathogenesis and carcinogenesis of LP. Further analysis of the EGFR family signaling pathway would clarify the pre-malignant nature of LP, and ErbB4 and its ligands might be possible new targets for the treatment of LP.
References
Silverman S Jr, Gorsky M, Lozada F . Oral leukoplakia and malignant transformation. A follow-up study of 257 patients. Cancer 1984; 53( 3): 563–568.
Warnakulasuriya S, Reibel J, Bouquot J et al. Oral epithelial dysplasia classification systems: predictive value, utility, weaknesses and scope for improvement. J Oral Pathol Med 2008; 37( 3): 127–133.
Reibel J . Prognosis of oral pre-malignant lesions: significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med 2003; 14( 1): 47–62.
Carpenter G, Cohen S . Epidermal growth factor. J Biol Chem 1990; 265( 14): 7709–7712.
Hirsch FR, Varella-Garcia M, Bunn PA Jr et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003; 21( 20): 3798–3807.
Morrison LE, Jacobson KK, Friedman M et al. Aberrant EGFR and chromosome 7 associate with outcome in laryngeal cancer. Laryngoscope 2005; 115( 7): 1212–1218.
Nicholson RI, Gee JM, Harper ME . EGFR and cancer prognosis. Eur J Cancer 2001; 37( Suppl 4): S9–S15.
Xia W, Lau YK, Zhang HZ et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res 1999; 5( 12): 4164–4174.
Shirasuna K, Hayashido Y, Sugiyama M et al. Immunohistochemical localization of epidermal growth factor (EGF) and EGF receptor in human oral mucosa and its malignancy. Virchows Arch A Pathol Anat Histopathol 1991; 418( 4): 349–353.
Revillion F, Lhotellier V, Hornez L et al. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann Oncol 2008; 19( 1): 73–80.
Ekberg T, Nestor M, Engstrom M et al. Expression of EGFR, HER2, HER3, and HER4 in metastatic squamous cell carcinomas of the oral cavity and base of tongue. Int J Oncol 2005; 26( 5): 1177–1185.
Del Sordo R, Angiero F, Bellezza G et al. HER family receptors expression in squamous cell carcinoma of the tongue: study of the possible prognostic and biological significance. J Oral Pathol Med 2010; 39( 1): 79–86.
Rautava J, Jee KJ, Miettinen PJ et al. ERBB receptors in developing, dysplastic and malignant oral epithelia. Oral Oncol 2008; 44( 3): 227–235.
Rubin Grandis J, Tweardy DJ, Melhem MF . Asynchronous modulation of transforming growth factor alpha and epidermal growth factor receptor protein expression in progression of premalignant lesions to head and neck squamous cell carcinoma. Clin Cancer Res 1998; 4( 1): 13–20.
Kimple RJ, Vaseva AV, Cox AD et al. Radiosensitization of epidermal growth factor receptor/HER2-positive pancreatic cancer is mediated by inhibition of Akt independent of ras mutational status. Clin Cancer Res 2010; 16( 3): 912–923.
Shigeishi H, Higashikawa K, Hiraoka M et al. Expression of epiregulin, a novel epidermal growth factor ligand associated with prognosis in human oral squamous cell carcinomas. Oncol Rep 2008; 19( 6): 1557–1564.
Marshall C, Blackburn E, Clark M et al. Neuregulins 1–4 are expressed in the cytoplasm or nuclei of ductal carcinoma (in situ) of the human breast. Breast Cancer Res Treat 2006; 96( 2): 163–168.
Kumagai K, Horikawa T, Gotoh A et al. Up-regulation of EGF receptor and its ligands, AREG, EREG, and HB-EGF in oral lichen planus. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010; 110( 6): 748–754.
Ulanovski D, Stern Y, Roizman P et al. Expression of EGFR and Cerb-B2 as prognostic factors in cancer of the tongue. Oral Oncol 2004; 40( 5): 532–537.
Bei R, Budillon A, Masuelli L et al. Frequent overexpression of multiple ErbB receptors by head and neck squamous cell carcinoma contrasts with rare antibody immunity in patients. J Pathol 2004; 204( 3): 317–325.
Bei R, Pompa G, Vitolo D et al. Co-localization of multiple ErbB receptors in stratified epithelium of oral squamous cell carcinoma. J Pathol 2001; 195( 3): 343–348.
Kramer IR, Lucas RB, el-Labban N et al. The use of discriminant analysis for examining the histological feathers of oral keratoses and lichen planus. Br J Cancer 1970; 24( 4): 673–683.
Maatta JA, Sundvall M, Junttila TT et al. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol Biol Cell 2006; 17( 1): 67–79.
Acknowledgements
We thank Dr Motohiro Kobayakawa, Dr Kazutoshi Kamei and Dr Naoki Hasegawa from Yokohama Rosai Hospital, and Dr Kazutoshi Nakaoka from Tsurumi University for providing some of the biopsy specimens used in this study. We also thank Applied Medical Research Laboratory for immunohistochemistry. This study was supported by a Grant-in-Aid for Young Scientists B 23792391 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kobayashi, H., Kumagai, K., Gotoh, A. et al. Upregulation of epidermal growth factor receptor 4 in oral leukoplakia. Int J Oral Sci 5, 14–20 (2013). https://doi.org/10.1038/ijos.2013.10
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijos.2013.10
Keywords
This article is cited by
-
Immunohistochemical expression of epidermal growth factor receptor (EGFR) in South Asian head and neck squamous cell carcinoma: association with various risk factors and clinico-pathologic and prognostic parameters
World Journal of Surgical Oncology (2018)
-
Targeted next-generation sequencing supports epidermoid metaplasia of the esophagus as a precursor to esophageal squamous neoplasia
Modern Pathology (2017)
-
Ado-trastuzumab emtansine-associated telangiectasias in metastatic breast cancer: a case series
Breast Cancer Research and Treatment (2014)