Abstract
Objectives:
The development of effective strategies to prevent childhood obesity and its comorbidities requires new, reliable early biomarkers. Here, we aimed to identify in peripheral blood cells potential transcript-based biomarkers of unhealthy metabolic profile associated to overweight/obesity in children.
Methods:
We performed a whole-genome microarray analysis in blood cells to identify genes differentially expressed between overweight and normal weight children to obtain novel transcript-based biomarkers predictive of metabolic complications.
Results:
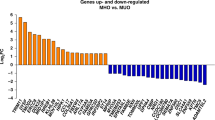
The most significant enriched pathway of differentially expressed genes was related to oxidative phosphorylation, for which most of genes were downregulated in overweight versus normal weight children. Other genes were involved in carbohydrate metabolism/glucose homoeostasis or in lipid metabolism (for example, TCF7L2, ADRB3, LIPE, GIPR), revealing plausible mechanisms according to existing biological knowledge. A set of differentially expressed genes was identified to discriminate in overweight children those with high or low triglyceride levels.
Conclusions:
Functional microarray analysis has revealed a set of potential blood-cell transcript-based biomarkers that may be a useful approach for early identification of children with higher predisposition to obesity-related metabolic alterations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
WHO. Obesity and overweight, 2015. Available at: http://www.who.int/mediacentre/factsheets/fs311/en/.
Ahrens W, Moreno LA, Marild S, Molnar D, Siani A, De Henauw S et al. Metabolic syndrome in young children: definitions and results of the IDEFICS study. Int J Obes 2014; 38 (Suppl 2): S4–S14.
Choquet H, Meyre D . Genomic insights into early-onset obesity. Genome Med 2010; 2: 36.
Bittel DC, Kibiryeva N, Sell SM, Strong TV, Butler MG . Whole genome microarray analysis of gene expression in Prader-Willi syndrome. Am J Med Genet A 2007; 143A: 430–442.
Silbiger VN, Luchessi AD, Hirata RD, Lima-Neto LG, Cavichioli D, Carracedo A et al. Novel genes detected by transcriptional profiling from whole-blood cells in patients with early onset of acute coronary syndrome. Clin Chim Acta 2013; 421: 184–190.
Visvikis-Siest S, Marteau JB, Samara A, Berrahmoune H, Marie B, Pfister M . Peripheral blood mononuclear cells (PBMCs): a possible model for studying cardiovascular biology systems. Clin Chem Lab Med 2007; 45: 1154–1168.
Sanchez J, Bonet ML, Keijer J, van Schothorst EM, Molller I, Chetrit C et al. Blood cells transcriptomics as source of potential biomarkers of articular health improvement: effects of oral intake of a rooster combs extract rich in hyaluronic acid. Genes Nutr 2014; 9: 417.
Caimari A, Oliver P, Keijer J, Palou A . Peripheral blood mononuclear cells as a model to study the response of energy homeostasis-related genes to acute changes in feeding conditions. Omics 2010; 14: 129–141.
Oliver P, Reynes B, Caimari A, Palou A . Peripheral blood mononuclear cells: a potential source of homeostatic imbalance markers associated with obesity development. Pflugers Arch 2013; 465: 459–468.
de Mello VD, Kolehmanien M, Schwab U, Pulkkinen L, Uusitupa M . Gene expression of peripheral blood mononuclear cells as a tool in dietary intervention studies: what do we know so far? Mol Nutr Food Res 2012; 56: 1160–1172.
Caimari A, Oliver P, Rodenburg W, Keijer J, Palou A . Feeding conditions control the expression of genes involved in sterol metabolism in peripheral blood mononuclear cells of normoweight and diet-induced (cafeteria) obese rats. J Nutr Biochem 2010; 21: 1127–1133.
Leonardson AS, Zhu J, Chen Y, Wang K, Lamb JR, Reitman M et al. The effect of food intake on gene expression in human peripheral blood. Hum Mol Genet 2010; 19: 159–169.
Sanchez J, Priego T, Pico C, Ahrens W, De Henauw S, Fraterman A et al. Blood cells as a source of transcriptional biomarkers of childhood obesity and its related metabolic alterations: results of the IDEFICS study. J Clin Endocrinol Metabol 2012; 97: E648–E652.
Konieczna J, Sanchez J, Palou M, Pico C, Palou A . Blood cell transcriptomic-based early biomarkers of adverse programming effects of gestational calorie restriction and their reversibility by leptin supplementation. Sci Rep 2015; 5: 9088.
Konieczna J, Sanchez J, van Schothorst EM, Torrens JM, Bunschoten A, Palou M et al. Identification of early transcriptome-based biomarkers related to lipid metabolism in peripheral blood mononuclear cells of rats nutritionally programmed for improved metabolic health. Genes Nutr 2014; 9: 366.
Ahrens W, Bammann K, Siani A, Buchecker K, De Henauw S, Iacoviello L et al. The IDEFICS cohort: design, characteristics and participation in the baseline survey. Int J Obes 2011; 35 (Suppl 1): S3–S15.
Cole TJ, Bellizzi MC, Flegal KM, Dietz WH . Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ 2000; 320: 1240–1243.
Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res 2010; 38 (Web Server issue): W210–W213.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I . Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001; 125: 279–284.
Cole TJ, Lobstein T . Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr Obes 2012; 7: 284–294.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003; 34: 267–273.
Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 2003; 100: 8466–8471.
Dahlman I, Forsgren M, Sjogren A, Nordstrom EA, Kaaman M, Naslund E et al. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-alpha. Diabetes 2006; 55: 1792–1799.
Takamura T, Honda M, Sakai Y, Ando H, Shimizu A, Ota T et al. Gene expression profiles in peripheral blood mononuclear cells reflect the pathophysiology of type 2 diabetes. Biochem Biophys Res Commun 2007; 361: 379–384.
Zhou Y, Park SY, Su J, Bailey K, Ottosson-Laakso E, Shcherbina L et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet 2014; 23: 6419–6431.
Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med 2007; 85: 777–782.
Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006; 38: 320–323.
Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 2006; 55: 2890–2895.
Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Investig 2007; 117: 2155–2163.
Garagnani P, Giuliani C, Pirazzini C, Olivieri F, Bacalini MG, Ostan R et al. Centenarians as super-controls to assess the biological relevance of genetic risk factors for common age-related diseases: a proof of principle on type 2 diabetes. Aging 2013; 5: 373–385.
Perez-Enciso M, Tenenhaus M . Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum Genet 2003; 112: 581–592.
Erhardt E, Czako M, Csernus K, Molnar D, Kosztolanyi G . The frequency of Trp64Arg polymorphism of the beta3-adrenergic receptor gene in healthy and obese Hungarian children and its association with cardiovascular risk factors. Eur J Clin Nutr 2005; 59: 955–959.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24.
Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 41: 25–34.
Pais R, Gribble FM, Reimann F . Stimulation of incretin secreting cells. Ther Adv Endocrinol Metab 2016; 7: 24–42.
McClean PL, Irwin N, Cassidy RS, Holst JJ, Gault VA, Flatt PR . GIP receptor antagonism reverses obesity, insulin resistance, and associated metabolic disturbances induced in mice by prolonged consumption of high-fat diet. Am J Physiol Endocrinol Metab 2007; 293: E1746–E1755.
Miyawaki K, Yamada Y, Ban N, Ihara Y, Tsukiyama K, Zhou H et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med 2002; 8: 738–742.
Gogebakan O, Osterhoff MA, Schuler R, Pivovarova O, Kruse M, Seltmann AC et al. GIP increases adipose tissue expression and blood levels of MCP-1 in humans and links high energy diets to inflammation: a randomised trial. Diabetologia 2015; 58: 1759–1768.
Lee AW, Hengstler H, Schwald K, Berriel-Diaz M, Loreth D, Kirsch M et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One 2012; 7: e41537.
Chanclon B, Luque RM, Cordoba-Chacon J, Gahete MD, Pozo-Salas AI, Castano JP et al. Role of endogenous cortistatin in the regulation of ghrelin system expression at pancreatic level under normal and obese conditions. PLoS One 2013; 8: e57834.
Rafacho A, Castellano-Muñoz M, Alonso-Magdalena P, Irles E, Bello M, Vetorazzi J et al. Cortistatin hyperpolarizes pancreatic beta cell membrane and reduces glucose-stimulated insulin secretion. Diabetol Metab Syndr 2015; 7 (Suppl 1): A250–A.
Acknowledgements
We are grateful for the support provided by school boards, headmasters, teachers, school staff and communities, and for the effort of all study nurses and data managers. This work was done as part of the IDEFICS study (http://www.idefics.eu) and the I.Family Study (http://www.ifamilystudy.eu/). We gratefully acknowledge the financial support of the European Community within the Sixth RTD Framework Programme Contract No. 016181 (FOOD) for the IDEFICS study and within the Seventh RTD Framework Programme Contract No. 266044 for the I.Family study. This work was also supported by the Instituto de Salud Carlos III, Centro de Investigación Biomédica en Red Fisiopatología de la Obesidad y Nutrición, CIBERobn. LBNB is a member of the European Research Network of Excellence NuGO (The European Nutrigenomics Organization, EU Contract: no. FP6-506360).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on International Journal of Obesity website
Supplementary information
Rights and permissions
About this article
Cite this article
Sánchez, J., Picó, C., Ahrens, W. et al. Transcriptome analysis in blood cells from children reveals potential early biomarkers of metabolic alterations. Int J Obes 41, 1481–1488 (2017). https://doi.org/10.1038/ijo.2017.132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2017.132
This article is cited by
-
KLB and NOX4 expression levels as potential blood-based transcriptional biomarkers of physical activity in children
Scientific Reports (2023)
-
Tejaas: reverse regression increases power for detecting trans-eQTLs
Genome Biology (2021)
-
Effect of black ginseng and silkworm supplementation on obesity, the transcriptome, and the gut microbiome of diet-induced overweight dogs
Scientific Reports (2021)
-
Distinct whole-blood transcriptome profile of children with metabolic healthy overweight/obesity compared to metabolic unhealthy overweight/obesity
Pediatric Research (2021)
-
Dietary intake of bioactive ingredients impacts liver and adipose tissue transcriptomes in a porcine model of prepubertal early obesity
Scientific Reports (2020)