Abstract
Background:
In rodents, hypothalamic brain-derived neurotrophic factor (BDNF) expression appears to be regulated by melanocortin-4 receptor (MC4R) activity. The impact of MC4R genetic variation on circulating BDNF in humans is unknown.
Objective:
The objective of this study is to compare BDNF concentrations of subjects with loss-of-function (LOF) and gain-of-function (GOF) MC4R variants with those of controls with common sequence MC4R.
Methods:
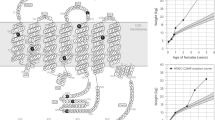
Circulating BDNF was measured in two cohorts with known MC4R sequence: 148 subjects of Pima Indian heritage ((mean±s.d.): age, 15.7±6.5 years; body mass index z-scores (BMI-Z), 1.63±1.03) and 69 subjects of Hispanic heritage (10.8±3.6 years; BMI-Z, 1.57±1.07). MC4R variants were characterized in vitro by cell surface expression, receptor binding and cyclic AMP response after agonist administration. BDNF single-nucleotide polymorphisms (SNPs) rs12291186, rs6265 and rs7124442 were also genotyped.
Results:
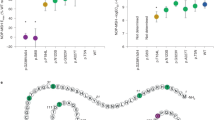
In the Pima cohort, no significant differences in serum BDNF was observed for 43 LOF subjects versus 65 LOF-matched controls (age, sex and BMI matched; P=0.29) or 20 GOF subjects versus 20 GOF-matched controls (P=0.40). Serum BDNF was significantly associated with genotype for BDNF rs12291186 (P=0.006) and rs6265 (P=0.009), but not rs7124442 (P=0.99); BDNF SNPs did not interact with MC4R status to predict serum BDNF. In the Hispanic cohort, plasma BDNF was not significantly different among 21 LOF subjects, 20 GOF subjects and 28 controls (P=0.79); plasma BDNF was not predicted by BDNF genotype or BDNF-x-MC4R genotype interaction.
Conclusions:
Circulating BDNF concentrations were not significantly associated with MC4R functional status, suggesting that peripheral BDNF does not directly reflect hypothalamic BDNF secretion and/or that MC4R signaling is not a significant regulator of the bulk of BDNF expression in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Xu B, Goulding EH, Zang K, Cepoi D, Cone RD, Jones KR et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci 2003; 6: 736–742.
Nicholson JR, Peter JC, Lecourt AC, Barde YA, Hofbauer KG . Melanocortin-4 receptor activation stimulates hypothalamic brain-derived neurotrophic factor release to regulate food intake, body temperature and cardiovascular function. J Neuroendocrinol 2007; 19: 974–982.
Bariohay B, Roux J, Tardivel C, Trouslard J, Jean A, Lebrun B . Brain-derived neurotrophic factor/tropomyosin-related kinase receptor type B signaling is a downstream effector of the brainstem melanocortin system in food intake control. Endocrinology 2009; 150: 2646–2653.
Caruso C, Carniglia L, Durand D, Gonzalez PV, Scimonelli TN, Lasaga M . Melanocortin 4 receptor activation induces brain-derived neurotrophic factor expression in rat astrocytes through cyclic AMP-protein kinase A pathway. Mol Cell Endocrinol 2013; 348: 47–54.
Kernie SG, Liebl DJ, Parada LF . BDNF regulates eating behavior and locomotor activity in mice. EMBO J 2000; 19: 1290–1300.
Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH et al. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci USA 1999; 96: 15239–15244.
Unger TJ, Calderon GA, Bradley LC, Sena-Esteves M, Rios M . Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. J Neurosci 2007; 27: 14265–14274.
Han JC, Liu QR, Jones M, Levinn RL, Menzie CM, Jefferson-George KS et al. Brain-derived neurotrophic factor and obesity in the WAGR syndrome. N Engl J Med 2008; 359: 918–927.
Gray J, Yeo GS, Cox JJ, Morton J, Adlam AL, Keogh JM et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes 2006; 55: 3366–3371.
Han JC, Muehlbauer MJ, Cui HN, Newgard CB, Haqq AM . Lower brain-derived neurotrophic factor in patients with prader-willi syndrome compared to obese and lean control subjects. J Clin Endocrinol Metab 2010; 95: 3532–3536.
Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci 2003; 23: 7143–7154.
Stranahan AM, Arumugam TV, Mattson MP . Lowering corticosterone levels reinstates hippocampal brain-derived neurotropic factor and Trkb expression without influencing deficits in hypothalamic brain-derived neurotropic factor expression in leptin receptor-deficient mice. Neuroendocrinology 2011; 93: 58–64.
Nakagawa T, Ono-Kishino M, Sugaru E, Yamanaka M, Taiji M, Noguchi H . Brain-derived neurotrophic factor (BDNF) regulates glucose and energy metabolism in diabetic mice. Diabetes Metab Res Rev 2002; 18: 185–191.
Bagnol D, Lu XY, Kaelin CB, Day HE, Ollmann M, Gantz I et al. Anatomy of qan endogenous antagonist: relationship between Agouti-related protein and proopiomelanocortin in brain. J Neurosci 1999; 19: RC26.
Farooqi IS, Yeo GS, Keogh JM, Aminian S, Jebb SA, Butler G et al. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J Clin Invest 2000; 106: 271–279.
Thearle MS, Muller YL, Hanson RL, Mullins M, Abdussamad M, Tran J et al. Greater impact of melanocortin-4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes 2012; 61: 250–257.
Xiang Z, Litherland SA, Sorensen NB, Proneth B, Wood MS, Shaw AM et al. Pharmacological characterization of 40 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists and the agouti-related protein (AGRP) antagonist. Biochemistry 2006; 45: 7277–7288.
Stutzmann F, Vatin V, Cauchi S, Morandi A, Jouret B, Landt O et al. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum Mol Genet 2007; 16: 1837–1844.
Krabbe KS, Nielsen AR, Krogh-Madsen R, Plomgaard P, Rasmussen P, Erikstrup C et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia 2007; 50: 431–438.
Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24.
Knowler WC, Pettitt DJ, Saad MF, Charles MA, Nelson RG, Howard BV et al. Obesity in the Pima Indians: its magnitude and relationship with diabetes. Am J Clin Nutr 1991; 53 (6 Suppl): 1543S–1551S.
Butte NF, Cai G, Cole SA, Comuzzie AG . Viva la Familia Study: genetic and environmental contributions to childhood obesity and its comorbidities in the Hispanic population. Am J Clin Nutr 2006; 84: 646–654.
Cole SA, Butte NF, Voruganti VS, Cai G, Haack K, Kent JW Jr et al. Evidence that multiple genetic variants of MC4R play a functional role in the regulation of energy expenditure and appetite in Hispanic children. Am J Clin Nutr 2010; 91: 191–199.
Tao YX . The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev 2010; 31: 506–543.
Cai G, Cole SA, Butte N, Bacino C, Diego V, Tan K et al. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity (Silver Spring) 2006; 14: 1596–1604.
Tan K, Pogozheva ID, Yeo GS, Hadaschik D, Keogh JM, Haskell-Leuvano C et al. Functional characterization and structural modeling of obesity associated mutations in the melanocortin 4 receptor. Endocrinology 2009; 150: 114–125.
Tao YX, Segaloff DL . Functional analyses of melanocortin-4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J Clin Endocrinol Metab 2005; 90: 5632–5638.
Xiang Z, Proneth B, Dirain ML, Litherland SA, Haskell-Luevano C . Pharmacological characterization of 30 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists, synthetic agonists, and the endogenous agouti-related protein antagonist. Biochemistry 2010; 49: 4583–4600.
Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB et al. 4-Norleucine, 7-D-phenylalanine-alpha-melanocyte-stimulating hormone: a highly potent alpha-melanotropin with ultralong biological activity. Proc Natl Acad Sci USA 1980; 77: 5754–5758.
Tao YX, Segaloff DL . Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology 2003; 144: 4544–4551.
Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 2012; 7: e51954.
Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z et al. 2000 CDC Growth Charts for the United States: methods and development. Vital Health Stat 2002; 11: 1–190.
Cole TJ . The LMS method for constructing normalized growth standards. Eur J Clin Nutr 1990; 44: 45–60.
Gaunt TR, Rodriguez S, Day IN . Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool 'CubeX'. BMC Bioinformatics 2007; 8: 428.
Katoh-Semba R, Wakako R, Komori T, Shigemi H, Miyazaki N, Ito H et al. Age-related changes in BDNF protein levels in human serum: differences between autism cases and normal controls. Int J Dev Neurosci 2007; 25: 367–372.
Webster MJ, Herman MM, Kleinman JE, Shannon Weickert C . BDNF and trkB mRNA expression in the hippocampus and temporal cortex during the human lifespan. Gene Expr Patterns 2006; 6: 941–951.
Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003; 112: 257–269.
Fujimura H, Altar CA, Chen R, Nakamura T, Nakahashi T, Kambayashi J et al. Brain-derived neurotrophic factor is stored in human platelets and released by agonist stimulation. Thromb Haemost 2002; 87: 728–734.
El-Gharbawy AH, Adler-Wailes DC, Mirch MC, Theim KR, Ranzenhofer L, Tanofsky-Kraff M et al. Serum brain-derived neurotrophic factor concentrations in lean and overweight children and adolescents. J Clin Endocrinol Metab 2006; 91: 3548–3552.
Acknowledgements
Funding for this study was provided by the International Hyperphagia Conference Best Idea Grant from the Prader–Willi Syndrome Association (USA) (JCH and JAY), by the NIH Intramural Research Programs of NICHD and NIDDK, American Diabetes Association grant 1–12-BS212 (Y-XT), NIH (R01 DK59264 and R01 DK080457 (NFB), and C06 RR013556 and C06 RR017515) and USDA/ARS under Cooperative Agreement 58–6250–51000–037. JK and JAY are Commissioned Officers of the United States Public Health Service. Clinical Trials Registry: Subjects in the Pima cohort were enrolled in observational, non-interventional studies, registered as NCT00339482. Subjects in the Hispanic cohort were enrolled in observational, non-interventional studies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on International Journal of Obesity website
Supplementary information
Rights and permissions
About this article
Cite this article
Hohenadel, M., Thearle, M., Grice, B. et al. Brain-derived neurotrophic factor in human subjects with function-altering melanocortin-4 receptor variants. Int J Obes 38, 1068–1074 (2014). https://doi.org/10.1038/ijo.2013.221
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2013.221
Keywords
This article is cited by
-
Early-Onset Obesity Caused by Monogenic Disorders
Current Pediatrics Reports (2017)