Abstract
C-reactive protein (CRP), an acute phase reactant and marker of inflammation, has been shown to be associated with CRP genetic variants and incident hypertension, but it is unclear whether this link is causal. We therefore conducted a prospective, nested case–control study to examine the relationship between single-nucleotide polymorphisms (SNPs) within the CRP gene, circulating CRP levels and the development of hypertension. Plasma CRP levels and the genotypes of eight SNPs were determined in 2000 unrelated Shanghai residents, including 908 hypertensive individuals and 1092 normotensive individuals. Among the 1092 normotensives, 968 subjects were followed up for 2 years, during which 71 developed hypertension. Plasma CRP levels were independently associated with the development of hypertension in the follow-up study (odds ratio per quartile=1.64; 95% confidence interval: 1.18–2.26; P<0.001). The minor alleles of rs1130864 (P<0.001) and rs3093059 (P<0.001) were significantly associated with elevated CRP levels, and the minor alleles of rs1205, rs1800947 and rs2246469 (all P<0.001) were associated with decreased CRP levels. A haplotype-based analysis strengthened the results of single-locus analysis. However, none of the SNPs or haplotypes was significantly associated with blood pressure, incident hypertension or changes between baseline and follow-up blood pressure levels. Taken together, our findings demonstrated that plasma CRP levels were substantially associated with common genetic variants in the CRP gene and could predict the development of hypertension. However, the relationship between genotype and CRP levels was not associated with a change in hypertension risk.
Similar content being viewed by others
Introduction
Mounting evidence suggests that persistent low-grade inflammation may be involved in the initiation and development of hypertension, with the exertion of proinflammatory actions through several mediators, including adhesion molecules, chemokines, growth factors, heat shock proteins, endothelin-1 and angiotensin.1, 2 Markers of inflammation may reflect subclinical vascular inflammation and thus be useful diagnostic tools. C-reactive protein (CRP), an acute phase reactant, is typically considered an inflammatory marker. Increased circulating concentrations of high-sensitivity CRP (hsCRP) have been associated with a panel of clinical endpoints, such as obesity, diabetes mellitus,3, 4, 5 hypertension2, 6, 7, 8 and atherosclerotic cardiovascular disease.9, 10, 11, 12, 13 The reported heritability estimates of CRP range from 27 to 40%. Common genetic variants in the CRP gene were found to be associated with plasma hsCRP levels,11, 13, 14, 15, 16 and this association may vary in different ethnic groups.17
As essential hypertension, accounting for 95% of cases of hypertension, is thought to arise from a mosaic network including a host of genetic and environmental factors that interact with each other,18, 19 it has been hypothesized that genetic variation in the CRP gene may influence plasma CRP levels and the subsequent development of hypertension. However, few prospective studies have investigated the causal relationship of plasma CRP and hypertension, especially in Chinese populations. Using existing sequence data and available haplotype-tagging SNPs, we comprehensively examined the common variations in the CRP gene in association with plasma CRP levels and tested whether these common genetic variants were associated with incident hypertension in a population-based prospective cohort of Chinese individuals.
Methods
Participants
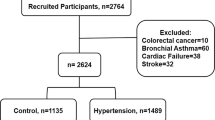
All subjects in this study were of Han Chinese descent and are currently residing in the Shanghai area. A random sample of 2000 unrelated subjects aged ⩾35 years were recruited in 2008, including 908 hypertensive patients and 1092 normotensive controls. The study protocol was reviewed and approved by the Ethics Committee of Shanghai Ruijin Hospital, and written informed consent was obtained from each subject. Of the 1092 normotensive controls with complete CRP levels and DNA samples, 968 were followed up after 2 years (in 2010), at which point 71 had developed hypertension.
Demographic information, including age, sex, smoking, drug treatment and cardiovascular disease history, was obtained from each patient using a standard questionnaire. Blood pressure was measured on three occasions with at least 10-min intervals by experienced examiners with the patients in a seated position according to a standard protocol recommended by the American Heart Association. All of the hypertensive patients met the following criteria: onset of hypertension before age 60 years; mean systolic blood pressure (SBP) ⩾140 mm Hg and/or mean diastolic blood pressure (DBP) ⩾90 mm Hg or the current use of antihypertensive drugs; and free of secondary causes of hypertension as assessed by extensive biochemical and clinical examination.
SNP selection and genotyping
We implemented a multistage approach to choosing haplotype-tagging single-nucleotide polymorphisms (tSNPs). First, three SNPs (rs1205, rs1800947 and rs3093059) were selected from the International HapMap Project collection of Han Chinese data,20 which were sufficient to assess the variation from 30 kb 5′ upstream to 30 kb 3′ downstream of the gene, with r2⩾0.9 and minor allele frequency ⩾5%, respectively. Second, another three SNPs (rs2211321, rs2027469 and rs12031749) from the NCBI dbSNP database were selected because their minor allele frequencies were ⩾5% in Asians. In addition, rs1130864 was included because it has been reported to be associated with CRP level.21, 22
Genomic DNA was extracted from leukocytes in a peripheral blood sample using a standard phenol–chloroform method. TaqMan SNP allelic discrimination was used as the primary genotyping technique. The genotyping protocol was performed on an ABI 7900HT real-time PCR machine (Applied Biosystems, Foster City, CA, USA) with primers and probes designed by Applied Biosystems. The genotypes were scored with SDS 2.2.2 allelic-differentiation software (Applied Biosystems). Approximately 5% of the samples were randomly selected to evaluate the reproducibility of the assay, and the concordance rate was more than 99%.
Measurement of biochemical variables
We collected fasting blood samples from each participant and processed the samples into separate serum, plasma and buffy coat aliquots. Baseline hsCRP was measured in plasma samples from 2000 subjects using an immunoturbidimetric assay on a Hitachi 911 analyzer (Roche Diagnostics, Chicago, IL, USA). The plasma triglyceride, total cholesterol, high-density lipoprotein cholesterol and fasting glucose concentrations in both the baseline and follow-up studies were determined enzymatically using commercially available kits and an autoanalyzer at the Shanghai Ruijin Hospital. Levels of low-density lipoprotein cholesterol were estimated using the Friedewald equation.
Statistical analysis
The mean values of continuous variables were compared using the Student’s t-test. The χ2 test or Fisher’s exact test was used to assess Hardy–Weinberg (H–W) equilibrium and to evaluate the differences in genotype/allele distributions between the two groups at baseline.
We log-transformed hsCRP concentrations with markedly skewed distributions to achieve near normal distributions and then calculated the differences in geometric mean values. To evaluate the association between CRP SNPs and plasma levels of CRP, we employed multivariable linear regression analyses that treated plasma hsCRP concentration as a dependent variable and tSNPs as independent variables, and controlled for factors such as age, sex, body mass index and cigarette smoking among the normotensive group. The odds ratio (OR) derived from the logistic regression was used to estimate the hazard ratio for the relationship between CRP and hypertension. Lewontin’s D prime (D′) and the correlation coefficient (r2) were calculated as two measures of linkage disequilibrium between CRP polymorphisms among normotensive participants. We employed the haplo.score and haplo.glm functions implemented in the Haplo.stats program to calculate haplotype frequencies and test for association with blood pressure and CRP levels. The analysis was adjusted for multiple testing using the false discovery rate method.23 The study power was calculated using PS (Power and Sample Size Calculations, Vanderbilt University, Nashville, TN, USA) software version 3.0.
Results
SNP data and clinical characteristics of the study population
All studied polymorphisms met the Hardy–Weinberg equilibrium in the normotensive group. Table 1 shows the data for the eight tag SNPs in CRP gene. There were no significant differences between hypertensives and normotensives for any of the studied SNPs at baseline, except for the genotypic distribution of rs3093059 (P=0.022). The clinical characteristics of the 2000 subjects analyzed in 2008 and the 968 subjects in the follow-up study in 2010 are presented in Table 2. Hypertensive patients had significantly higher age, body mass index, triglycerides, fasting glucose and CRP levels but lower HDL cholesterol levels than normotensive controls, and they were more likely to be men.
Association of plasma CRP with blood pressure and hypertension
As shown in Table 3, the cross-sectional analysis indicated that plasma CRP levels increased with both SBP and DBP (P<0.001). The prevalence of hypertension increased with plasma CRP levels after adjusting for confounding factors (OR per quartile=1.39; 95% confidence interval (CI): 1.22–1.58; P<0.001).
Among 968 normotensive subjects, the incidence of hypertension measured in 2010 was positively correlated with the plasma CRP levels measured in 2008 (OR=1.64; 95% CI: 1.18–2.26; P<0.001). CRP levels were significantly associated with changes in SBP (P=0.002).
Association of SNPs with plasma CRP levels and hypertension
Among 1092 normotensive controls at baseline, 5 out of the 8 tSNPs genotyped were significantly associated with plasma hsCRP levels (Figure 1). The minor alleles of rs1130864 and rs3093059 were significantly associated with 32.1 and 22.3% higher hsCRP levels, respectively. In contrast, the minor alleles of rs1205, rs1800947 and rs2246469 were associated with 24–38% decreases in plasma CRP levels. Among all 2000 subjects, none of the SNPs was significantly associated with prevalent hypertension after adjusting for age and sex (Supplementary Table 1). Similar results were obtained for quantitative blood pressure traits when subjects on anti-hypertensive medication were adjusted by SBP/DBP of 10/5 mm Hg (Supplementary Table 2).
Association between CRP genetic variants and CRP plasma concentrations (mg l−1) among 1092 normotensive subjects in additive genetic effect model. The error bar shows the 95% CI of the geometric mean. The x axis shows the genotypes for each SNP in the order of homozygous major alleles, heterzygotes and homozygous minor alleles. The P-value were adjusted for age and sex.
Association of haplotypes with plasma CRP levels, risk of hypertension and blood pressure
Haplotypes were constructed using the five SNPs (rs1205, rs1130864, rs3093059, rs180094 and rs2246469) that were associated with CRP levels. Five common haplotypes (H1–H5) had an estimated frequency ⩾5% (Table 4). Four haplotypes were associated with plasma CRP levels, similar to the association of individual SNPs with CRP levels. Haplotypes H1 and H5, which contained the minor alleles of rs1130864 and rs3093059 and the major allele of rs1205, were associated with elevated baseline plasma CRP levels (Hap-scores=2.05 and 2.35, respectively, P<0.05), and H4, which included the minor alleles of rs1205 and rs180094, was associated with decreased CRP levels (Hap-score=−2.87, P=0.004). All haplotypes had a nonsignificant association with blood pressure, prevalence of hypertension and changes in blood pressure (all P>0.05).
Discussion
In this study, we investigated the relationship between plasma CRP levels and the risk of developing hypertension using a prospective, nested case–control study design. Plasma CRP levels were found to be significantly associated with both SBP and DBP, and an elevated CRP level at baseline was predictive of the development of hypertension. Our study, conducted in a cohort with a wide age range, is consistent with the previous findings. Among female US health professionals aged ⩾45 years24 and elderly subjects aged ⩾65 years,8 CRP levels were associated with the future development of hypertension, suggesting that inflammation contributes to the pathogenesis of hypertension.
Some animal studies supported the results of our study. For example, Guan et al.25 found that transgenic mice with CRP overexpression exhibited efficient and sustained hCRP expression and increased blood pressure. This effect was associated with the increased expression of the angiotensin type 1 receptor, serum endothelin 1 and the endothelin type A receptor, and the decreased expression of the angiotensin type 2 receptor and endothelial NO synthase in the thoracic aorta. Nagai et al.26 showed that transgenic transverse aortic constriction mice overexpressing the human CRP gene had higher myocardial messenger RNA levels of interleukin-6, CD68 and collagen-I, and greater levels of infiltrating Mac-2-positive macrophages and reactive oxygen species than nontransgenic × transverse aortic constriction mice. At 4 weeks after surgery, the heart and lung weights and extent of cardiac fibrosis were greater in transgenic transverse aortic constriction mice. Therefore, CRP itself may have a pathogenic role in the development of pressure overload-induced cardiac remodeling, possibly through enhanced inflammation and oxidative stress.
Although studies that assessed common variation and haplotypes in the CRP gene with plasma CRP levels have used different SNP selection and haplotype estimation methodologies, our study confirmed previous reports of the association of plasma CRP with genetic variants, especially that the minor alleles of rs1205 and rs1800947 are associated with lower CRP levels and that the minor allele of rs1130864 is associated with higher CRP levels.12, 15, 16, 17, 21 Moreover, we found that the G allele of rs2246469 was associated with lower CRP levels in Han Chinese.
The use of CRP genetic variants as instrumental variables has been widely applied to study whether CRP has a direct pathogenic role in vascular diseases or is just an ‘innocent bystander’. Thus far, the results of these studies have been controversial. As low-grade inflammation has been associated with endothelial damage, hypertension, atherosclerosis and other components of metabolic syndrome, we analyzed all of these diseases in a comprehensive way. On the one hand, in a large German sample,21 three SNPs (rs3093075, rs1130864 and rs1800947) showed a significant association with microangiopathic stroke. In Turkish adults,27 four SNPs (rs3091244, rs1130864, rs1800947 and rs1205) were studied, and the haplotypes CGCA and TGTG were associated with elevated serum CRP levels and hypertension in women and men, respectively. On the other hand, Lawlor et al.28 reported no associations between CHD and a genetic variant (rs1130864) that is known to be related to CRP levels, and the Rotterdam Study11 reported that four haplotypes significantly associated with CRP levels were not associated with CHD. Similarly, in healthy elderly Japanese,29 rs3091244, rs1130864 and the T–T–G (rs1341665, rs3091244 and rs1800947) haplotype were identified as genetic markers for elevated basal CRP levels, but rs1800947 and the C–C–C haplotype appeared to be correlated with arterial pulse wave velocity.
In this study, we failed to find any significant association between SNPs in CRP and hypertension; that is to say, the relationship between genotype and CRP levels largely did not translate to an associated change in hypertension risk. It remains possible that the elevation in CRP levels could be caused by environmental and lifestyle factors such as smoking, obesity and insulin resistance.30 In addition to environmental stimuli, CRP is induced by other inflammatory cytokines, and additional genes may regulate CRP levels as well.31 As stated by Danesh et al.,32 although elevated CRP was a predictor of cardiovascular disease, it added little to current risk prediction models based on established traditional risk factors.
Several limitations of this study merit consideration. First, the 2-year follow-up period is somewhat short, and only 71 subjects developed hypertension during this time. Owing to the small sample size of this study, we could not exclude the possibility of an association between CRP SNPs and hypertension/blood pressure. However, we had more than 85% statistical power to detect an OR of 1.64 for hypertension in the follow-up cohort. Moreover, this community-based cohort is relatively socioeconomically homogeneous and may not represent a random sample of the Chinese Han population. Nevertheless, we were able to analyze the relationship between CRP and hypertension using both cross-sectional and prospective study designs in the same study. In addition, the SNPs we selected capture most of the variations in the CRP gene, and our haplotype-based analysis was consistent with the single-SNP–plasma hsCRP associations in both direction and magnitude.
In conclusion, our study confirms that plasma CRP levels predict the development of hypertension in Han Chinese. The steady-state plasma CRP levels are influenced by CRP SNPs and haplotypes, but our study does not support the hypothesis that common variations in the CRP gene exert a substantial effect on hypertension risk. Further studies are warranted to confirm the ethnicity-related differences in CRP gene variants and the potential direct roles of these genetic variants on the risk of hypertension.
References
Bautista LE . Inflammation, endothelial dysfunction, and the risk of high blood pressure: epidemiologic and biological evidence. J Hum Hypertens 2003; 17: 223–230.
Boos CJ, Lip GY . Elevated high-sensitive C-reactive protein, large arterial stiffness and atherosclerosis: a relationship between inflammation and hypertension? J Hum Hypertens 2005; 19: 511–513.
Hu FB, Meigs JB, Li TY, Rifai N, Manson JE . Inflammatory markers and risk of developing type 2 diabetes in women. Diabetes 2004; 53: 693–700.
Liu S, Tinker L, Song Y, Rifai N, Bonds DE, Cook NR, Heiss G, Howard BV, Hotamisligil GS, Hu FB, Kuller LH, Manson JE . A prospective study of inflammatory cytokines and diabetes mellitus in a multiethnic cohort of postmenopausal women. Arch Intern Med 2007; 167: 1676–1685.
Zee RY, Germer S, Thomas A, Raji A, Rhees B, Ridker PM, Lindpaintner K, Williams GH, Nathan DM, Martin M . C-reactive protein gene variation and type 2 diabetes mellitus: a case-control study. Atherosclerosis 2008; 197: 931–936.
Hommels MJ, van der Ven AJ, Kroon AA, Kessels AG, van Dieijen-Visser MP, van Engelshoven JA, Bruggeman CA, de Leeuw PW . C-reactive protein, atherosclerosis and kidney function in hypertensive patients. J Hum Hypertens 2005; 19: 521–526.
Davey Smith G, Lawlor DA, Harbord R, Timpson N, Rumley A, Lowe GD, Day IN, Ebrahim S . Association of C-reactive protein with blood pressure and hypertension: life course confounding and mendelian randomization tests of causality. Arterioscler Thromb Vasc Biol 2005; 25: 1051–1056.
Dauphinot V, Roche F, Kossovsky MP, Schott AM, Pichot V, Gaspoz JM, Gosse P, Barthelemy JC . C-reactive protein implications in new-onset hypertension in a healthy population initially aged 65 years: the Proof study. J Hypertens 2009; 27: 736–743.
Ridker PM, Hennekens CH, Buring JE, Rifai N . C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med 2000; 342: 836–843.
Pai JK, Pischon T, Ma J, Manson JE, Hankinson SE, Joshipura K, Curhan GC, Rifai N, Cannuscio CC, Stampfer MJ, Rimm EB . Inflammatory markers and the risk of coronary heart disease in men and women. N Engl J Med 2004; 351: 2599–2610.
Kardys I, de Maat MP, Uitterlinden AG, Hofman A, Witteman JC . C-reactive protein gene haplotypes and risk of coronary heart disease: the Rotterdam Study. Eur Heart J 2006; 27: 1331–1337.
Miller DT, Zee RY, Suk Danik J, Kozlowski P, Chasman DI, Lazarus R, Cook NR, Ridker PM, Kwiatkowski DJ . Association of common CRP gene variants with CRP levels and cardiovascular events. Ann Hum Genet 2005; 69 (Part 6): 623–638.
Pai JK, Mukamal KJ, Rexrode KM, Rimm EB . C-reactive protein (CRP) gene polymorphisms, CRP levels, and risk of incident coronary heart disease in two nested case-control studies. PLoS One 2008; 3: e1395.
Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, Tracy RP . Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis 2001; 154: 681–689.
Suk Danik J, Chasman DI, Cannon CP, Miller DT, Zee RY, Kozlowski P, Kwiatkowski DJ, Ridker PM . Influence of genetic variation in the C-reactive protein gene on the inflammatory response during and after acute coronary ischemia. Ann Hum Genet 2006; 70 (Part 6): 705–716.
Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, Witrak L, Rieder MJ, Nickerson DA . Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation 2006; 114: 2458–2465.
Lee CC, You NC, Song Y, Hsu YH, Manson J, Nathan L, Tinker L, Liu S . Relation of genetic variation in the gene coding for C-reactive protein with its plasma protein concentrations: findings from the Women’s Health Initiative Observational Cohort. Clin Chem 2009; 55: 351–360.
Lifton RP, Gharavi AG, Geller DS . Molecular mechanisms of human hypertension. Cell 2001; 104: 545–556.
Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel JM . A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature 1992; 355: 262–265.
International HapMap Consortium A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Kuhlenbaeumer G, Huge A, Berger K, Kessler C, Voelzke H, Funke H, Stoegbauer F, Stoll M, Ringelstein EB . Genetic variants in the C-reactive protein gene are associated with microangiopathic ischemic stroke. Cerebrovasc Dis 2010; 30: 476–482.
Portoles J, Del Peso G, Fernandez-Reyes MJ, Bajo MA, Lopez-Sanchez P . Previous comorbidity and lack of patient free choice of technique predict early mortality in peritoneal dialysis. Perit Dial Int 2009; 29: 150–157.
Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, Lumley T, Bis JC, Wiggins KL, Rosendaal FR, Psaty BM . Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA 2007; 297: 489–498.
Sesso HD, Buring JE, Rifai N, Blake GJ, Gaziano JM, Ridker PM . C-reactive protein and the risk of developing hypertension. JAMA 2003; 290: 2945–2951.
Guan H, Wang P, Hui R, Edin ML, Zeldin DC, Wang DW . Adeno-associated virus-mediated human C-reactive protein gene delivery causes endothelial dysfunction and hypertension in rats. Clin Chem 2009; 55: 274–284.
Nagai T, Anzai T, Kaneko H, Mano Y, Anzai A, Maekawa Y, Takahashi T, Meguro T, Yoshikawa T, Fukuda K . C-reactive protein overexpression exacerbates pressure overload-induced cardiac remodeling through enhanced inflammatory response. Hypertension 2011; 57: 208–215.
Komurcu-Bayrak E, Erginel-Unaltuna N, Onat A, Ozsait B, Eklund C, Hurme M, Mononen N, Laaksonen R, Hergenc G, Lehtimaki T . Association of C-reactive protein (CRP) gene allelic variants with serum CRP levels and hypertension in Turkish adults. Atherosclerosis 2009; 206: 474–479.
Lawlor DA, Harbord RM, Timpson NJ, Lowe GD, Rumley A, Gaunt TR, Baker I, Yarnell JW, Kivimaki M, Kumari M, Norman PE, Jamrozik K, Hankey GJ, Almeida OP, Flicker L, Warrington N, Marmot MG, Ben-Shlomo Y, Palmer LJ, Day IN, Ebrahim S, Smith GD . The association of C-reactive protein and CRP genotype with coronary heart disease: findings from five studies with 4,610 cases amongst 18,637 participants. PLoS One 2008; 3: e3011.
Morita A, Nakayama T, Doba N, Hinohara S, Soma M . Polymorphism of the C-reactive protein (CRP) gene is related to serum CRP level and arterial pulse wave velocity in healthy elderly Japanese. Hypertens Res 2006; 29: 323–331.
Hak AE, Stehouwer CD, Bots ML, Polderman KH, Schalkwijk CG, Westendorp IC, Hofman A, Witteman JC . Associations of C-reactive protein with measures of obesity, insulin resistance, and subclinical atherosclerosis in healthy, middle-aged women. Arterioscler Thromb Vasc Biol 1999; 19: 1986–1991.
Hubacek JA, Peasey A, Pikhart H, Stavek P, Kubinova R, Marmot M, Bobak M . APOE polymorphism and its effect on plasma C-reactive protein levels in a large general population sample. Hum Immunol 2010; 71: 304–308.
Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GD, Pepys MB, Gudnason V . C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med 2004; 350: 1387–1397.
Acknowledgements
This study was supported by project 973 from the Ministry of Science and Technology of China (grant 2009cB521905), the Shanghai Hospital Development Center (grant 2007315) and the Shanghai Commission of Science and Technology (grant 08DZ2200400 and 08410701900). We gratefully acknowledge the assistance and cooperation of the faculty and staff of the Department of Hypertension at Ruijin Hospital, as well as the study participants for their enthusiastic participation.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on Hypertension Research website
Supplementary information
Rights and permissions
About this article
Cite this article
Kong, H., Qian, YS., Tang, XF. et al. C-reactive protein (CRP) gene polymorphisms, CRP levels and risk of incident essential hypertension: findings from an observational cohort of Han Chinese. Hypertens Res 35, 1019–1023 (2012). https://doi.org/10.1038/hr.2012.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hr.2012.89
Keywords
This article is cited by
-
Association between CRP genetic diversity and bipolar disorder comorbid complications
International Journal of Bipolar Disorders (2018)
-
GWAS-Supported CRP Gene Polymorphisms and Functional Outcome of Large Artery Atherosclerotic Stroke in Han Chinese
NeuroMolecular Medicine (2018)
-
C-Reactive Protein Gene Contributes to the Genetic Susceptibility of Hemorrhagic Stroke in Men: a Case-Control Study in Chinese Han Population
Journal of Molecular Neuroscience (2017)
-
Genetic polymorphisms in C-reactive protein increase cancer susceptibility
Scientific Reports (2016)
-
C-reactive protein genetic variant is associated with diabetic retinopathy in Chinese patients with type 2 diabetes
BMC Endocrine Disorders (2015)