Abstract
Anthocyanins are secondary metabolites in land plants that contribute to the colors of leaves and flowers, and are nutritionally valuable components of the human diet. The DFR gene plays an important role in the anthocyanin biosynthetic pathway. In this study, we investigated the regulation of DFR expression and in different Malus crabapple cultivars that show distinct patterns of leaf coloration, and how it influences leaf anthocyanin accumulation and coloration. Specifically, we studied the ever-red leaved cultivar ‘Royalty’, the ever-green leaved cultivar ‘Flame’ and the spring-red leaved cultivar ‘Radiant’. RT-PCR analysis showed that the expression of McDFR1 correlated with the expression of a MYB transcription factor, McMYB10, and with anthocyanin accumulation. We isolated five McDFR1 promoter fragments from the three cultivars and identified four different fragments (F1–4) that were present either in several cultivars, or only in one. Yeast one-hybrid and electrophoretic mobility shift assay analyses showed that McMYB10 could bind to all the McDFR1 promoters, except McDFR1-Ra2. The F1, F2 and F3 fragments did not affect McMYB10 binding to the McDFR1 promoters; however, we found evidence that the F4 fragment suppressed binding, and that the MYBGAHV amino-acid sequence maybe an important cis-element for McMYB10 protein binding. This information has potential value for strategies to modify plant color through genetic transformation.
Similar content being viewed by others
Introduction
Anthocyanins comprise a class of water-soluble flavonoids pigments in plants that contribute to the color of flowers, fruit, stems and leaves.1,2 They also function in vegetative tissues that provide protection against UV and high light irradiation, as antioxidants to scavenge reactive oxygen species (ROS), and as antimicrobial agents during defense responses.3–6 Anthocyanins are also potentially beneficial components of the human diet,6,7 and they can act as antioxidants, are anti-carcinogenic,8,9 anti-inflammatory10 and may help support both diabetes prevention and treatment,11 and heart health.12
Over the past few decades, the anthocyanin biosynthetic pathway has been well characterized in plants, such as arabidopsis (Arabidopsis thaliana), petunia (Petunia hybrida), maize (Zea mays), snapdragon (Antirrhinum majus) and apple (Malus domestica).13–17 Most of the genes encoding enzymes responsible for each step in the flavonoid biosynthetic pathway have been identified,18 and it has been established that dihydroflavonol 4-reductase (DFR) plays a key role in the formation of common and condensed anthocyanins (Figure 1).18 DFR is one of the rate-limiting enzymes that catalyzes the production of flavan-3,4-diols (leucoanthocyanidins) via the reduction of three colorless dihydroflavonols: dihydrokaempferol (DHK), dihydroquercetin (DHQ); and dihydromyricetin (DHM). These three compounds are also intermediates in flavonol biosynthesis through the flavonol synthase reaction, using NADPH as a cofactor.19,20
The flavonoid pathway leading to the synthesis of anthocyanins and flavonols. ANS, anthocyanidin synthase; CHI, chalcone isomerase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; DFR, dihydroflavonol reductase; F3H, flavonoid 3-hydroxylase; PAL, phenylalanine ammonia lyase; 4CL, 4-coumarate CoA ligase.
A number of DFR genes have been characterized from a wide range of plant species, including Camellia sinensis (tea),21 Medicago truncatula (Medicago),22 Ipomoea batatas Lam (sweet potato),23 Ginkgo biloba (ginkgo)24 and Populus trichocarpa.25 It has also been shown that overexpression of different DFR genes in tobacco (Nicotiana benthamiana) flowers promotes anthocyanin biosynthesis, corresponding to an increase in red pigmentation.26 In addition, suppressing IbDFR in sweet potato led to a decrease in anthocyanin accumulation and reduced the tolerance to abiotic stress.23 DFR not only regulates levels of anthocyanin, but also shows substrate specificity, resulting in the accumulation of different types of anthocyanins. For example, petunia (Petunia hybrida (J. D. Hooker) Vilmorin) and cymbidium (Cymbidium faberi Rolfe) lack varieties with brick red/orange colored flowers due to a lack of pelargonidin-based anthocyanins because DFRs do not utilize dihydrokaempferol as a substrate.27,28 Furthermore, overexpressing MtDFR in rice also changed several metabolites except anthocyanins, including the concentrations of amino acids, sugars and metals.29 Altered the expression of DFR also affect the expression of other anthocyanin biosynthesis gene. The upregulation of McDFR expression in crabapple leaves or apple peels was accompanied by a proportional increase in some of the genes involved in anthocyanin biosynthesis (McCHS, McCHI, McF3H, McF3’H, McDFR, McANS and McUFGT).30 In addition, overexpression of RrDFR1 (Rosa rugosa) and PhDFR1 (Petunia hybrida) genes in tobacco displayed downregulation of the endogenous NtFLS gene, and the promotion of anthocyanin synthesis. The relative expression levels of NtCHS and NtFLS were significantly downregulated and NtANS genes was upregulated in DFR transgenic lines.26
In plants, MYB (v-myb avian myeloblastosis viral oncogene homolog), bHLH (basic-helix-loop-helix) and WD40 (Trp-Asp 40) transcription factors (TFs) often form MYB-bHLH-WD40 (MBW) complexes and play important roles in secondary metabolism, development, signal transduction and disease resistance. It has been shown that several MYB TFs are involved in positively regulating the expression of DFR.31 In A. thaliana, the expression of the MYB75/PAP1 gene is induced by light and precedes the expression of both MYB90/PAP2 and several structural genes (CHS, DFR, F3H and LDOX).32 Moreover, the apple TFs MdMYB10 and MdMYB1 control the red pigmentation in the fruit flesh and skin, respectively,33 and the corresponding proteins, MdMYB10 and MdMYB1, can form complexes with bHLH genes to trans-activate the DFR promoter and promote anthocyanin accumulation.34 miR156 targets, SPL9, negatively regulates anthocyanin accumulation by directly preventing expression of anthocyanin biosynthetic genes DFR through destabilization of a MYB-bHLH-WD40 (PAP1, TT8 and TTG1) transcriptional activation complex.32 In contrast, several MYB transcription factors also have been proved that they can suppress the expression of DFR. AtMYBL2 interacts with TT8 (TRANSPARENT TESTA 8) to reduce anthocyanin biosynthesis by suppressing the expression of DFR.33 The expression of the AtMYB7 gene is induced by salt stress, and represses several flavonoid pathway genes, including DFR and UGT.35 Furthermore, MYBCORE, MYBGAHV cis- regulatory elements have been proved that involved in MYB transcription factors binding to several promoters to regulate gene expression, such as MYB101 regulates fertilization in Arabidopsis thaliana by binding to the MYBGAHV elements in the promoters of downstream genes, and MYB5 and TT2 regulates proanthocyanins accumulation in Arabidopsis seed coat by binding to the MYBCORE elements in the promoter of DFR.36,37
Structural differences in promoter regions contribute to the difference in expression of the target genes regulated by TFs.38 In apple, a TATA-box insertion in the IRT1 promoter is responsible for increasing Fe uptake, and its presence correlates with an increase in transcriptional activation by specific binding of the TF, IID.39 Meanwhile, promoter structure also affect anthocyanins-related gene expression in plants. Subsequent analysis showed that a rearrangement of the 23-bp sequences in the promoter regions of MdMYB10 and McMYB10 was responsible for the difference in gene expression between the white- and red-fleshed apples and red-leaf color crabapples.33 Specifically, the R1: MdMYB10 (McMYB10) promoter has a single MdMYB10 (McMYB10) binding motif, and is only present in white-fleshed apples, while the R6: MdMYB10 (McMYB10) promoter, which is present in red-fleshed apples, has five additional tandem repeats of the MdMYB10 (McMYB10) binding motif. Our studies in Malus crabapple have shown that a 743-bp fragment missing of McCHS promoter was found in ever-green leaf color crabapple cultivar, and the missing sequence contained two MYBPLANT elements and three MYBCORE elements that are essential for MYB transcription factor binding and inducing anthocyanin biosynthesis. So, we conclude that different structures in the chalcone synthase (McCHS) promoter affect the binding of MYB TFs and the expression of McCHS, and lead to the accumulation of different anthocyanins in crabapple cultivars with different leaf colors.40 However, little is known about the basis for the differences in DFR expression caused by MYB TFs.
Malus crabapples are collectively an economically important germplasm resource for ornamental plants, providing numerous landscape species. They also exhibit stress resistance and provide valuable research material to investigate the mechanisms of anthocyanin accumulation and color formation, reflected by the diversity of leaf, flower and fruit coloration.41 In this current study, the expression analysis showed that McDFR1 may play an important role in anthocyanins biosynthesis in crabapple leaves, hence we examined differences in the McDFR1 promoter sequences and functions in three different types of Malus crabapple: Malus cv. ‘Flame’, Malus cv. ‘Royalty’ and Malus cv. ‘Radiant’. We investigated whether variations in the DFR1 promoter may affect the expression of DFR1 in different colored leaf crabapple cultivars and provide evidence that the McDFR1 promoter has potential applications to improve plant color via genetic transformation approaches.
Materials and methods
Material and growth conditions
Three crabapple cultivars were used in this study: ‘Royalty’, an ever-red leaved cultivar; ‘Flame’, an ever-green leaved cultivar; and ‘Radiant’, which has red young leaves that turn green as they mature. Eight-year-old crabapple trees grafted onto a Malus ‘Balenghaitang’ stock were grown in the Crabapple Germplasm Resources Nursery, Beijing University of Agriculture (40.l″ north latitude, 116.6″ longitude). In mid-late April (18–22 °C) and on sunny days, we selected six trees of each cultivar that showed similar growth and collected leaf samples from annual branches growing in all four compass directions on the outer edge of each canopy. Leaves of ‘Royalty’, ‘Radiant’ and ‘Flame’ were collected at 10 developmental stages (3, 6, 9, 12, 15, 18, 21, 24, 27 and 30 days after budding). Branches to be used for the experiments were marked before budding. All samples were immediately frozen in liquid nitrogen and stored at −80 °C.
High-performance liquid chromatography analysis
The crabapple leaves (approximately 0.8–1.0 g fresh weight) were extracted with 10 mL extraction solution (methanol:water:formic acid:trifluoroacetic acid=70:27:2:1),42 at 4 °C in the dark for 72 h, with shaking every 6 h. The homogenate was then filtered through sheets of qualitative filter paper and the filtrate was then passed through a 0.22 μL reinforced nylon membrane filter (Millipore, Billerica, MA, USA). The resulting sample was subjected to high-performance liquid chromatography (HPLC) using an HPLC1100-DAD system (Agilent Technologies, Waldbronn, Germany), with trifluoroacetic acid: formic acid: water (0.1:2:97.9) as mobile phase A and trifluoroacetic acid: formic acid: acetonitrile: water (0.1:2:48:49.9) as mobile phase B.43 The gradients used were as follows: 0 min, 30% B; 10 min, 40% B; 50 min, 55% B; 70 min, 60% B; 80 min, 30% B. Detection of anthocyanins was performed at 520 nm. All samples were analyzed in biological triplicate.
Cloning and analysis of the McDFR1 promoters
To analyze the differences in McDFR1 promoter sequences of the three different leaf colored crabapple cultivars, genomic DNA was isolated from ‘Royalty’, ‘Radiant’ and ‘Flame’ leaves using the Plant Genomic DNA Kit (TIANGEN BIOTECH CO., LTD, Beijing, China). The 5′-upstream sequences were amplified by hi-TAIL PCR44 the primers are shown in Supplementary Table S1. The primers for hi-TAIL PCR (high-efficient thermal asymmetric interlaced PCR) were designed based on the McDFRs cDNA sequence (GenBank Accession: FJ817487, AF117268). The upstream-flanking sequence of McDFR1 was isolated using hi-TAIL PCR. The hi-TAIL PCR is a method to isolate upstream (promoters) and downstream sequences of the known coding sequences. Long (33–34 nucleotides) arbitrary degenerate (LAD) primer with a higher degree (2304 or 6912 folds) of degeneracy is used to create primer-binding sites efficiently along the unknown target sequences and used for the first amplification. An additional sequence (AC1, 18 nucleotides with Tm=58 °C, used for the second and third amplification) identical to the 5′ part of the LAD primer is tagged to the 5′ end of a long primer (Tm>68 °C) specific to the known sequence. The forward special primers (McDFR1-promoter1, McDFR1-promoter2, and McDFR1-promoter3; Supplementary Table S1) used in three reactions were based on the McDFR1 coding sequence (GenBank Accession: FJ817487). All PCR products were sub-cloned into the pGEM T-Easy Vector (Promega, Madison, WI, USA) and transformed into Escherichia coli DH5α cells and sequenced. The cis-elements were analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/).
RNA extraction and semi-quantitative RT-PCR
Total RNA was extracted from crabapple leaves using the RNA Extract kit (Aidlab, Beijing, China) according to the manufacturer’s instructions. DNase I (TaKara, Ohtsu, Japan) was added to remove genomic DNA, and the 1 μg RNA samples were subjected to cDNA synthesis using the Access RT-PCR System (Promega), according to the manufacturer’s instructions.
Semi-quantitative RT-PCR was carried out in 20 μL reactions with 2 μL of 10× diluted cDNA template, and McActin was used as the internal control.45 transcription system (Promega) PCR amplification system with the following conditions: initial denaturation at 95 °C for 3 min, followed by 20 or 29 cycles (29 cycles for McDFR1, McDFR1, McMYB10 and 20 cycles for McActin) of 95 °C for 10 s, 60 °C (for McDFR1, McDFR1, McMYB10 and McActin) for 30 s and extension at 72 °C for 30 s and final denaturation was at 72 °C for 5 min, maintain at 4 °C. Specific primers for semi-quantitative RT-PCR analysis were designed using primer 5 software and are listed in Supplementary Table S1.46
The expression levels of McDFR1, McDFR2 and McMYB10 in crabapple were analyzed using qRT-PCR and the SYBR Green qPCR Mix (TaKaRa) and the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA), according to the manufacturers’ instructions. The PCR primers were designed using NCBI Primer BLAST and are listed in Supplementary Table S1. qRT-PCR analysis was carried out in a total volume of 20 μL containing 9 μL of 2×SYBR Green qPCR Mix (TaKaRa), 0.1 μM specific primers (each), and 100 ng of template cDNA. The reaction mixtures were heated to 95 °C for 30 s, followed by 39 cycles at 95 °C for 10 s, 50–59 °C for 15 s and 72 °C for 30 s. A melting curve was generated for each sample at the end of each run to ensure the purity of the amplified products. The transcript levels were normalized using the Malus 18S ribosomal RNA gene (DQ341382, for apple and crabapple) as the internal controls and calculated using the 2(−ΔΔCt) analysis method.46
Yeast one-hybrid assay
A yeast one-hybrid system was used to assay the relationship between the McMYB10 protein and the McDFR1 promoters from the three Malus crabapple cultivars.47 As the effector construct, the open reading frame of McMYB10 was cloned into the BamHI and SalI sites of the pJG4-5 vector (Clontech, Palo Alto, CA, USA) under the control of the galactokinase 1 (GAL1) promoter. The McDFR1 promoter sequences were inserted upstream of the LacZ reporter gene in the pLacZi vector. The effector and reporter or control constructs were transformed into competent cells of the yeast strain EGY48, resulting in the following yeast strains: pJG4-5-McMYB10/ pLacZi-promoters of McDFR1; pJG4-5/ pLacZi-promoters of McDFR1; pJG4-5-McMYB10/pLacZi; and pJG4-5/ pLacZi. The yeast cells were selected on synthetic drop-out media lacking tryptophan and uracil, and positive colonies were spotted onto glucose plates (2%) containing X-gal at 28 °C for 2 days and examined for blue color development.45
Electrophoretic mobility shift assay
The McMYB10 open reading frame sequence was cloned into the pMAL-C2X expression vector and transformed into Rosetta (DE3) E. coli competent cells.48 The maltose-binding protein (MBP)-tag encoded in the pMAL-C2X vector was used to facilitate purification of the recombinant protein. Isopropyl β-d-1-thiogalactopyranoside (0.3 mM) was added to the cultures to induce McMYB10 expression in a cell culture shaking at 170 r.p.m. for 6 h at 28 °C. The recombinant protein was purified with the One-Stop MBP-Tagged Protein Miniprep Pack (BioLab. Co. Ltd, Beijing, China). Electrophoretic mobility shift assay (EMSA) reactions were prepared according to the manufacturer’s protocol (LightShif Chemi luminescent EMSA Kit; Termo Fisher Scientifc, Waltham, MA, USA). The probes were labeled by annealing biotin-labeled oligonucleotides. The same 100×unlabeled DNA fragment was used as a competitor in the assay. The oligonucleotides used for EMSA are listed in Supplementary Table S2. Approximately 10 μg of purified McMYB10 recombinant protein was used for each EMSA reaction.
Results
McDFR expression and anthocyanin accumulation during leaf development
McDFR is known to play an important role in anthocyanin biosynthesis.49 There are two DFR homologs in crabapple, and the full-length McDFR cDNAs of these two genes were cloned from cDNA libraries that from the leaf of ever-red leaf color crabapple cultivar ‘Royalty’, and named McDFR1 and McDFR2 (FJ817487, AF117268). To verify the relationship between the expression levels of the McDFR genes and anthocyanin accumulation, we examined anthocyanin accumulation and the expression profiles of the McDFR genes and McMYB10 in 10 leaf developmental stages (3, 6, 9, 12, 15, 18, 21, 24, 27 and 30 days after budding) of the three crabapple cultivars by HPLC and RT-PCR, respectively (Figure 2).
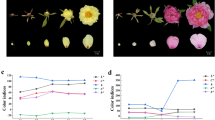
Analysis of flavonoid accumulation and the expression profiles of McDFR genes and McMYB10 during 10 leaf developmental stages in the Malus crabapple ever-red cultivar ‘Royalty’, spring-red cultivar ‘Radiant’ and ever-green cultivar ‘Flame’. (a) The anthocyanin content in 10 leaf developmental stages of ‘Royalty’, ‘Radiant’ and ‘Flame’. (b) The expression of McDFR1, McDFR2 and McMYB10 analyzed by RT-PCR. (c) Real-time PCR was used to analyze McDFR1, McDFR2, McMYB10 expression patterns in the leaves of ‘Royalty’, ‘Radiant’ and ‘Flame’. Malus 18S (DQ341382) was used as the reference gene. Error bars indicate the s.e.m.±s.e. of three replicate measurements. Different letters above the bars indicate significantly different values (P<0.05) calculated using one-way analysis of variance (ANOVA) followed by a Tukey’s multiple range test. ND, no detection. 1–10 represents stage 1–10 of leaf development stages. 1, 3 days after budding; 2, 6 days after budding; 3, 9 days after budding; 4, 12 days after budding; 5, 15 days after budding; 6, 18 days after budding; 7, 24 days after budding; 8, 21 days after budding; 9, 27 days after budding; 10, 30 days after budding.
We observed that in the ever-red ‘Royalty’ cultivar, the abundance of anthocyanins was relatively high in the first six developmental stages, before decreasing to low levels in the last four stages. Anthocyanin levels increased during the first five development stages and a gradual decrease was observed in the last five development stages in the spring-red crabapple cultivar ‘Radiant’, while they were almost undetectable in the ever-green ‘Flame’ cultivar.
RT-PCR analyses showed that the expression level of McDFR1 were higher expressed in the first four and seven stages and gradually decreased in ‘Royalty’ and ‘Radiant’, respectively, but gradually increase in ‘Flame’ and highest in the stage of eight. The transcription of McDFR2 gradually decrease in these three crabapple cultivars except stage 9 in ‘Radiant’. McMYB10 was not detected in ‘Flame’ leaves and showed almost same expression trend with McDFR1 in ‘Royalty’ and ‘Radiant’. We also performed qRT-PCR to quantified the expression distinction of McDFR1, McDFR2 and McMYB10 in these three cultivars, the results further confirmed that expression variations of these three genes in three crabapple cultivars. The expression level of McDFR1 and McMYB10 in leaf colored cultivars significantly higher than that in green-leaf crabapple cultivar ‘Flame’. Furthermore, the higher expression level of McDFR2 in ‘Flame’ was observed, from which we inferred that McDFR2 maybe involved in the biosynthesis of other flavonoid compounds than anthocyanins. Meanwhile, the correlation analysis showed the expression of McDFR1 is closely related to the accumulation of anthocyanins and the transcription of McMYB10 in crabapple leaves, and McDFR2 only have 25.3% and 8.8% relativity with anthocyanins accumulation in ‘Royalty’ and ‘Radiant’ during leaf development, respectively (Supplementary Table S3).
Taken together, these data suggest that McDFR1 and McMYB10 expression is associated with anthocyanin accumulation in crabapple leaves and that McMYB10 may be involved in the modulation of anthocyanin levels by regulating McDFR1. However, the mechanism by which McMYB10 controls the expression of McDFR1 is unclear.
Sequence analysis of the McDFR1 promoter and differences between the three Malus crabapple cultivars
Since the expression level of McDFR1 is consistent with anthocyanins accumulation and the transcription of McMYB10, we speculated that McDFR1 may play an important role in anthocyanins biosynthesis in crabapple leaves, hence we focused on the reason underlying this expression difference of McDFR1 in ‘Royalty’, ‘Radiant’ and ‘Flame’. We demonstrate there are natural variations in the promoter region of the McDFR1 gene. McDFR1 promoter from ‘Royalty’ is homozygous (1590 bp, MF592793), named McDFR1-R; McDFR1 promoters from ‘Flame’ are heterozygous, alleles of the ‘Flame’ McDFR1 promoters were named McDFR1-F1 and McDFR1-F2 (F1, 1603 bp, MF592792; F2, 1445 bp, KT276932.1), respectively; McDFR1 promoters from ‘Radiant’ are also heterozygous, McDFR1 promoters were named McDFR1-Ra1 and McDFR1-Ra2 (Ra1, 1610 bp, MF592794; Ra2, 1693 bp, MF592795), respectively.
Among the five McDFR1 promoter sequences, we found four different sequences that were either shared, or unique, and named them F1, F2, F3 and F4 (Figure 3). The F1 fragment (160 bp) was detected in the McDFR1-F1, McDFR1-R and McDFR1-Ra1 promoters, the F2 fragment (28 bp) was found in the McDFR1-F2 and McDFR1-Ra2 promoters, and the F3 fragment (19 bp), an AT-rich fragment, was found in the McDFR1-F1 and McDFR1-Ra1 promoters. Interestingly, a 234 bp insertion was only observed in the McDFR1-Ra2 promoter. We hypothesized that McDFR1 promoter sequence diversity may influence the expression levels of McDFR1.
The sequence characteristics of the McDFR1 promoter of ‘Flame’, ‘Royalty’ and ‘Radiant’. Alignment of the promoter sequences of McDFR1in three typical leaf colored crabapple cultivars. The McDFR1 promoter from ‘Royalty’ was named McDFR1-R, the two McDFR1 promoters from ‘Flame’ were named McDFR1-F1 and McDFR1-F2, and the two McDFR1 promoters from ‘Radiant’ were named McDFR1-Ra1 and McDFR1-Ra2. We named the different fragments F1, F2, F3 and F4, respectively.
Cis-element analysis of the McDFR1 promoter
To understand the mechanism of McDFR1 transcriptional regulation by MYB TFs, we used the homologous cis-regulatory elements of the five McDFR1 promoter sequences to search for known elements using the PLACE database,50,51 with a focus on putative MYB-binding sites. The MYB2AT, MYB2CONSENSUSAT, MYBATRD22, MYBCORE, MYBCOREATCYCB1 and MYBPZM elements, which are required for MYB TF binding, were present in almost the same positions in the five McDFR1 promoters (Table 1). However, the number and the positions of MYB1AT, MYB1LEPR, MYBGAHV and MYBST1 varied. MYB1AT and MYBST1 were located 800 bp upstream of the ATG transcriptional start site and were present in various numbers, while MYB1LEPR were present 600 bp upstream from the ATG in the two promoters that did not contain F1 fragments. MYBGAHV was present in the two ‘Flame’ McDFR1 promoters, the McDFR1-R promoter and the McDFR1-Ra1 promoter, but not in the McDFR1-Ra2 promoter, ~300 bp upstream of the ATG (Table 1 and Figure 4). We deduced that the MYBGAHV element may play a role in regulation of McDFR1 expression by MYB TFs.
Interaction of the McDFR1 promoter with McMYB10
To test the hypothesis that the F1, F2, F3 and F4 fragments affect the expression of McDFR1, which is regulated by McMYB10 in crabapple, we evaluated McMYB10 binding to the McDFR1-F1, McDFR1-F2, McDFR1-R, McDFR1-Ra1, McDFR1-Ra2 using the yeast one-hybrid assay (Figure 5a). LacZ activity was detected in yeast cells containing pJG4-5-McMYB10 together with pLacZi-proMcDFR1-F1, pLacZi-proMcDFR1-F2, pLacZi-proMcDFR1-R, pLacZi-proMcDFR1-Ra1, but not in yeast harboring pLacZi-proMcDFR1-Ra2 together with pJG4-5-McMYB10, or in the control. Hence we hypothesized that the absence of MYBGAHV element or existence of F4 fragment might affect the activity of McMYB10 binding to the McDFR1 promoter. To further investigate whether McMYB10 binds directly to the MYB1AT, MYB1LEPR, MYBGAHV and MYBST1 cis-elements, we performed an EMSA (Figure 5b). Biotin-labeled probes were designed to extend 10–20 bp on either side of the elements located nearest to the ATG transcriptional start site of each sequence. We found that McMYB10 bound to all four elements, and that this binding diminished gradually with 100× concentration of unlabeled probe. Furthermore, the binding of MYB1AT, MYBGAHV and MYBST1 to the McMYB10 protein was stronger than that of MYB1LEPR. Then to determine whether F4 fragment involved in McMYB10 binding to the McDFR1 promoter, we generated pLacZi-proMcDFR1-Ra2 (without F4), which were based on the native Ra2 promoter sequences, but without F4 fragment. The yeast cells containing pJG4-5-McMYB10 and pLacZi-proMcDFR1-Ra2 (without F4) showed a light blue color.
Cis-element binding ability and yeast one-hybrid assay of the McDFR1 promoters with McMYB10. (a) Interaction of the McMYB10 protein with the McDFR1-F1, McDFR1-F2, McDFR1-R, McDFR1-Ra1, McDFR1-Ra2 and McDFR1-Ra2 (without F4) promoter regions, as revealed using yeast one-hybrid assays. The panel shows yeast cells containing distinct effector and reporter constructs grown on an SD/-Trp/-Ura medium plate. The interaction of McMYB10, fused to the GAL4 activation domain (pJG4-5-McMYB10), with LacZ driven by McDFR promoters (pLacZi-promoters of McDFR1) is shown in the bottom panel. Yeast transformed with pJG4-5-McMYB10/pLacZi, pJG4-5/pLacZi-McDFR1 promoters and pJG4-5/pLacZi were used as controls. (b) Electrophoretic mobility shift assay (EMSA) of four the cis-elements, MYB1AT, MYBGAHV, MYB1LEPR, MYBST1, with McMYB10. 100×Unlabeled probe refers to the control of adding 100 times the concentration of a competing non-labeled specific probe. The black arrow indicates protein-DNA complexes, and the white arrow shows the positions of free probes. In lanes with competitor DNA, there was an excess of unlabeled probe.
From these results we concluded that the F4 fragment may suppress the binding of McMYB10 to the McDFR1-Ra2 promoter, and that the absence of MYBGAHV from the McDFR1-Ra2 promoter maybe the main reason why McMYB10 bound only weakly to the McDFR1-Ra2 (without F4) promoter. The F1, F2 and F3 McDFR1 promoter fragments did not affect McMYB10 binding to the McDFR1 promoters.
Discussion
There is considerable interest in the breeding of ornamental plants with colored leaves, especially those with red/purple leaves, which is mainly due to the accumulation of anthocyanins.52–54 However, the molecular mechanism of red color formation in leaves is still unclear. DFR is a key enzyme in the catalysis of the stereo-specific reduction of dihydroflavonols to leucoanthocyanidins, which uses NADPH as a cofactor and is located in a key branch point in the anthocyanin biosynthetic pathway.55 The expression level variations of DFR determine the color of leaves, flowers and fruits in many plants,56–58 and given the central role of DFR in anthocyanin accumulation, we investigated the possible regulatory mechanism by which MYB TFs control DFR gene expression in crabapple leaves.
HPLC analysis and transcript quantification suggested that red leaves were associated with higher anthocyanin accumulation, which was consistent with increased transcript levels of McDFR and McMYB10 (Figure 2). Several studies have shown that MYB10 can activate the expression of DFR and promote anthocyanin accumulation. In Arabidopsis thaliana, AtDFR is a target gene of MYB75/PAP1 and its expression can be enhanced by an elevated expression of MYB75.59 MdMYB10 and MdMYB1 have been shown to be able to bind the DFR promoter and promote anthocyanin accumulation in apple skin.34 Therefore, we hypothesized that McDFR maybe a McMYB10 target and that high expression of McDFR might result in red leaf coloration. To address this, we examined the expression level of two McDFR genes and McMYB10 in three crabapple cultivars and the results showed that McDFR1 have positively relationship with the expression of McMYB10 and anthocyanins accumulation in crabapple leaves. Hence, we focused on the regulation mechanism of McDFR1 in crabapple. We cloned five McDFR1 promoter fragments. Interestingly, low expression of the McDFR genes and low expression of McMYB10 were observed in the ever-green leaf colored cultivar ‘Flame’ (Figure 2a). We speculate that McDFR genes is responsible for the accumulation of flavonoid products other than anthocyanins in ‘Flame’. Moreover, since LacZ activity driven by the McDFR1-F1, McDFR1-F2 and McDFR1-R promoters in the yeast one-hybrid assay (Figure 5a) was detectable, we hypothesize that there was no anthocyanin accumulation in the green leaves as a consequence of low MYB10 expression, and the low level of expression of genes in the anthocyanin biosynthetic pathway.
In a previous study, 1 kb insertion in the promoter that increases the expression of citrate transporter gene HvAACT1 in several Al-tolerant barley cultivars.60 In addition, a deletion in the promoter of a mitochondrial molybdenum transporter gene (MOT1) in Arabidopsis is associated with reduced gene expression and low molybdenum (Mo) levels in the shoot.61 The same nucleotide polymorphisms were also appeared in anthocyanins-related genes. An extra fragment (255 bp) in the promoter of TfF3′H1 in reddish flower tulip sport Tulipa fosteriana hampered the accumulation of cyanidin anthocyanins through the reduction of TfF3′H1 transcription.62 A 23 bp repeat motif in the upstream regulatory region of MYB10 alleles was found only in red-fleshed apples and red leaved crabapples.33,63 Moreover, this promoter allele was shown to be responsible for the increased accumulation of anthocyanins, and the number of repeat units correlated with an increase in trans-activation by the MYB10 protein.33 A 743 bp deletion fragment in the McCHS gene promoter in the ever-green leaf colored crabapple cultivar ‘Flame’ would lead to only a few MYB TFs binding to the promoter sequence and reduce McCHS expression, reflecting a direct relationship with anthocyanin content and leaf color.33 This suggests structural differences in the promoter are an important factor in determining the gene expression variations in the different cultivars. To determine the expression pattern of McDFR1 in different crabapple cultivars, we isolated five McDFR1 promoter fragments from three typical leaf colored cultivars (Figure 3). The results showed that the promoters of McDFR1 contain many MYB-binding cis-elements, consistent with McDFR1 being regulated by MYB TFs. The yeast one-hybrid results showed that McMYB10 could not bind to the McDFR1-Ra2 promoter, but when the F4 fragment was deleted in the McDFR1-Ra2 promoter, binding was partially restored compared with the original McDFR1-Ra2 promoter (Figure 5a). Furthermore, EMSA analyses suggested that McMYB10 binds to the MYB1AT, MYBGAHV, MYB1LEPR and MYBST1 cis-elements in the McDFR1 promoters (Figure 5b), and we also found that the MYBGAHV element that was not present in McDFR1-Ra2 and McDFR1-Ra2 (without F4) affected McMYB10 binding to the DFR1 promoter to increase DFR expression. We concluded that the F4 fragment in the McDFR1-Ra2 promoter sequence determined whether or not MYB10 binds to the McDFR1 promoter, and that the MYBGAHV element may be an important cis-element in regulating DFR1 expression by MYB10. Meanwhile, we also analyzed the potential bHLH or WD40 binding sites in F4 fragment, the results showed that F4 fragment lack related transcription factors binding sites. Hence, we speculate that McDFR1-Ra1 promoter was preferentially used in ‘Radiant’ to make the leaf present red color in early development stages. And the McDFR1-Ra2 promoter was keep low activity in ‘Radiant’ during leaf development.
PCR analysis showed that both the R6 and R1 promoters of McMYB10 were present in the genome simultaneously in spring-red crabapple cultivars,33 and the same phenomenon (different promoters of one gene exist in one cultivars) was observed in the McDFR1 promoter in spring-red crabapple cultivars. The McDFR1-Ra2 promoter, which contains the important repressive F4 fragment and the McDFR1-Ra1 promoter, which is strongly bound by McMYB10, are both present in the spring-red crabapple cultivar ‘Radiant’. We therefore deduce that the key anthocyanin-related genes contain activated and non-activated promoters simultaneously, and this may be a characteristic of the genomes of cultivars where red and green leaf color co-exist.
Alteration of the expression of DFR in plants represents an excellent strategy to modify the content of anthocyanins and flavonoids. In this regard, a suitable promoter is critical for the expression of exogenous genes, and our data suggest that the F4 fragment and the MYBGAHV cis-element may be used as valuable tools for the regulation of gene expression.
References
Andersen M, Jordheim M . Anthocyanins. Encyclopedia of Life Sciences (ELS). John Wiley and Sons Ltd: Chichester: 2010; 1–12.
Gould K, Davies K, Winefield C . Anthocyanins: Biosynthesis, Functions, and Application. Springer: New York, NY, USA, 2009.
Koes R, Verweij W, Quattrocchio F . Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 2005; 10: 236–242.
Lepiniec L, Debeaujon I, Routaboul JM et al. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 2006; 57: 405–430.
Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I . Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 2007; 12: 29–36.
Mouradov A, Spangenberg G . Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front Plant Sci 2014; 5: 620.
Lila MA . Anthocyanins and human health: an in vitro investigative approach. J Biomed Biotechnol 2004; 5: 306–313.
Bagchi D, Sen C, Bagchi M, Atalay M . Anti-angiogenic, antioxidant, and anti-carcinogenic properties of a novel anthocyanin-rich berry extract formula. Biochemistry (Mosc) 2004; 69: 75–80.
Butelli E, Titta L, Giorgio M et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 2008; 26: 1301–1308.
Ohgami K, Ilieva I, Shiratori K et al. Anti-inflammatory effects of aronia extract on rat endotoxin-induced uveitis. Invest Ophthalmol Vis Sci 2005; 46: 275–281.
Tsuda T . Recent progress in anti-obesity and anti-diabetes effect of berries. Antioxidants 2016; 5: 13.
Mazza G . Anthocyanins and heart health. Dellistituto Superiore Di Sanita 2007; 43: 369–374.
Winkel-Shirley B . Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 2001; 126: 485–493.
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM . Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J 2008; 53: 814–827.
Pourcel L, Bohorquez-Restrepo A, Irani NG, Grotewold E ‘Anthocyanin biosynthesis, regulation, and transport: new insights from model species’. Recent Advances in Polyphenol Research, Wiley-Blackwell: New Jersey, NJ, 2012, pp 143–160.
Petroni K, Tonelli C . Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 2011; 181: 219–229.
Espley RV, Bava C, Bovy A et al. Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol J 2013; 11: 408–419.
Holton TA, Cornish EC . Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995; 7: 1071–1083.
Martens S, Knott J, Seitz CA, Janvari L, Yu SN, Forkmann G . Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissues. Biochem Eng J 2003; 14: 227–235.
Peters D, Constabel CP . Molecular analysis of herbivore-induced condensed tannin synthesis: cloning, expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J Cell Mol Biol 2002; 32: 701–712.
Singh K, Kumar S, Yadav SK, Ahuja PS . Characterization of dihydroflavonol 4-reductase cDNA in tea (Camellia sinensis (L.) O. Kuntze). Plant Biotechnol Rep 2009; 3: 95–101.
Xie DY, Jackson LA, Cooper JD, Ferreira D, Paiva NL . Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 2004; 134: 979–994.
Wang H, Fan W, Li H, Yang J, Huang J, Zhang P . Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 2013; 8: e78484.
Cheng H, Li L, Cheng S et al. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS ONE 2013; 8: e72017.
Huang Y, Gou J, Jia Z et al. Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS ONE 2012; 7: e30364.
Luo P, Ning G, Wang Z et al. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front Plant Sci 2015; 6: 1257.
Forkmann G, Heller W . ‘Biosynthesis of flavonoids. In:Sankawa, U (ed.). Polyketides and Other Secondary Metabolites Including Fatty Acids and their Derivatives. A. Elsevier: Amsterdam, AMS, Netherland, 1999, 713–748.
Johnson ET, Yi H, Shin B, Oh BJ, Cheong H, Choi G . Cymbidium hybrida dihydroflavonol-4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J Cell Mol Biol 1999; 19: 81–85.
Takahashi H, Hayashi M, Goto F et al. Evaluation of metabolic alteration in transgenic rice overexpressing dihydroflavonol 4-reductase. Ann Bot 2016; 98: 819–825.
Tian J, Han ZY, Zhang J, Hu Y, Song T, Yao Y . The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci Rep 2015; 5: 12228.
Cominelli E, Gusmaroli G, Allegra D et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 2008; 165: 886.
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW . Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2001; 23: 1512–1522.
Espley RV, Brendolise C, Chagné D et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 2009; 21: 168–183.
Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D . AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 2014; 55: 507–516.
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR . Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 2016; 142: 1216–1232.
Xu W, Grain D, Bobet S et al. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 2014; 202: 132–144.
Liang Y, Tan Z, Zhu L et al. MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLos Genet 2013; 9: e1003933.
Lin-Wang K, Bolitho K, Grafton K et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 2010; 10: 50.
Huang W, Lv H, Wang Y . Functional characterization of a novel R2R3-MYB transcription factor modulating the flavonoid biosynthetic pathway from Epimedium sagittatum. Front Plant Sci 2017; 8: 1274.
Tian J, Shen H, Zhang J, Song T, Yao Y . Characteristics of chalcone synthase promoters from different leaf-color Malus crabapple cultivars. Sci Hort 2011; 129: 449–458.
Wang YS, Gao LP, Shan Y, Liu YJ, Tian YW, Xia T . Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci Hort 2012; 141: 7–16.
Hashimoto F, Tanaka M, Maeda H, Shimizu K, Sakata Y . Characterization of cyanic flower color of Delphinium cultivars. Engei Gakkai Zasshi 2008; 69: 428–434.
Revilla E, Ryan JM . Analysis of several phenolic compounds with potential antioxidant properties in grape extracts and wines by high-performance liquid chromatography-photodiode array detection without sample preparation. J Chromatogr A 2000; 881: 461–469.
Liu YG, Chen Y . High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2017; 43: 649.
Tian J, Peng Z, Zhang J et al. McMYB10 regulates coloration via activating McF3’H and later structural genes in ever-red leaf crabapple. Plant Biotechnol J 2015; 13: 948–961.
Tai DQ, Tian J, Zhang J, Song TT, Yao YC . A Malus crabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS ONE 2014; 10: e110570.
Tripathi P, Prunedapaz JL, Kay SA . A modified yeast-one hybrid system for heteromeric protein complex-DNA interaction studies. J Vis Exp 2017; 24.
Tian J, Zhang J, Han ZY et al. McMYB12 transcription factors co-regulate proanthocyanidin and anthocyanin biosynthesis in Malus crabapple. Sci Rep 2017; 7: 43715.
Ahmed NU, Park JI, Jung HJ, Yang TJ, Hur Y, Nou IS . Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 2014; 550: 46–55.
Higo K, Ugawa Y, Iwamoto M, Korenaga T . Plant cis-acting regulatory DNA elements (PLACE) database. Nucelic Acids Res 1999; 27: 297–300.
Prestridge DS . SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Bioinformatics 1991; 7: 203–206.
Lightbourn GJ, Griesbach RJ, Novotny JA, Clevidence BA, Rao DD, Stommel JR . Effects of anthocyanin and carotenoid combinations on foliage and immature fruit color of Capsicum annuum L. Journal of Heredity 2008; 99: 105–111.
Lebowitz RJ. The genetics and breeding of coleus,’ In: Jules J (ed.). Plant Breeding Reviews, 1985; pp 343–360.
Nguyen P, Cin VD . The role of light on foliage colour development in coleus (Solenostemon scutellarioides (L.) Codd). Plant Physiol Biochem 2009; 47: 934–945.
Chu Y, Pan J, Wu A, Cai R, Chen H . Molecular cloning and functional characterization of dihydroflavonol-4-reductase, gene from calibrachoa hybrida. Sci Hort 2014; 165: 398–403.
Kim J, Lee W, Vu T, Jeong C, Hong S, Lee H . High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep 2017; 36: 1–10.
Li Y, Liu X, Cai X et al. Dihydroflavonol 4-reductase genes from Freesia hybrid play important and partially overlapping roles in the biosynthesis of flavonoids. Front Plant Sci 2017; 8: e72017.
Massonnet M, Fasoli M, Tornielli GB et al. Transcriptomic differences in grapevine varieties correlate with berry anthocyanin skin. Plant Physiol 2017 pp 00311.
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C . Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000; 12: 2383–2394.
Fujii M, Yokosho K, Yamaji N et al. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 2012; 3: 713.
Baxter I, Muthukumar B, Park HC et al. Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet 2008; 4: e1000004.
Yuan Y, Ma X, Tang D, Shi Y . Comparison of anthocyanin components, expression of anthocyanin biosynthetic structural genes, and TfF3’H1, sequences between Tulipa fosteriana, ‘Albert heijn’ and its reddish sport. Sci Hort 2014; 175: 16–26.
Murray R, Boase C, Brendolise L et al. Failure to launch: the self-regulating MdMYB10 R6, gene from apple is active in flowers but not leaves of petunia. Plant Cell Rep 2015; 34: 1817–1823.
Acknowledgements
Financial support was provided by the ‘National Natural Science Foundation of China’ (31501723), ‘The Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions’, the Beijing Technology Innovation Service Capacity Construction-Research Plan (KM201610020003), Scientific research improvement project of BUA (GL2015002) and ‘the Project of Construction of Innovative Teams and the Teacher Career Development for Universities and Colleges Under Beijing Municipality’ (IDHT20150503). We thank the Fruit Tree Key Laboratory at the Beijing University of Agriculture and the Beijing Nursery Engineering Research Center for Fruit Crops for providing experimental resources. We are grateful to all technicians in the BUA Crabapple Germplasm Resource Garden. We also thank PlantScribe (www.plantscribe.com) for editing this manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: YY. Performed the experiments: XZ, KL, TS. Analyzed the data: MC, KL, TS. Contributed reagents/materials/analysis tools: JT, YY, TS. Wrote the paper: JT, YY and JZ.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information for this article can be found on the Horticulture Research website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tian, J., Chen, Mc., Zhang, J. et al. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic Res 4, 17070 (2017). https://doi.org/10.1038/hortres.2017.70
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hortres.2017.70
This article is cited by
-
Chromosomal level genome assemblies of two Malus crabapple cultivars Flame and Royalty
Scientific Data (2024)
-
Integrated metabolome and transcriptome analysis unveils novel pathway involved in the fruit coloration of Nitraria tangutorum Bobr.
BMC Plant Biology (2023)
-
Molecular evolution and expression assessment of DFRs in apple
Chemical and Biological Technologies in Agriculture (2023)
-
Effects of editing DFR genes on flowers, leaves, and roots of tobacco
BMC Plant Biology (2023)
-
Extremophiles as Plant Probiotics to Promote Germination and Alleviate Salt Stress in Soybean
Journal of Plant Growth Regulation (2023)