Abstract
Mutations in KAT6A, encoding a member of the MYST family of histone acetyl-transferases, were recently reported in patients with a neurodevelopmental disorder (OMIM: #616268, autosomal dominant mental retardation-32). In this report, we describe three siblings with intellectual disability (ID) or global developmental delay and a KAT6A heterozygous nonsense mutation, i.e., c.3070C>T (p.R1024*, ENST00000406337; chr8:41795056G>A on hg19). This mutation was identified by whole-exome sequencing of all three siblings but not in a healthy sibling. The mutation was not detected in the peripheral blood of their parents, suggesting the existence of parental germline mosaicism. The primary symptoms of our patients included severe to profound ID or global developmental delay, including speech delay with craniofacial dysmorphism; these symptoms are consistent with symptoms previously described for patients with KAT6A mutations. Although several features are common among patients with KAT6A mutations, the features are relatively nonspecific, making it difficult to establish a clinical entity based on clinical findings alone. To the best of our knowledge, this is the first report of cases with a KAT6A mutation in an Asian population and these cases represent the first reported instances of germline mosaicism of this disease.
Similar content being viewed by others
Introduction
Intellectual disability (ID) is defined as an intelligence quotient of 70 or below and affects ~1% of children worldwide.1 ID is caused by environmental or genetic factors, but the explicit cause is not identified in up to 60% of cases.2 Approximately 25–50% of ID cases are thought to have a genetic cause.2 A recent large-scale sequencing study identified probable pathogenic mutations in ~40% individuals with ID who underwent whole-exome sequencing (WES) or genome sequencing.3
WES is a powerful tool in identifying genetic alterations in putative genetic disorders, even those that are undiagnosed, providing a molecular diagnosis rate of 25%.4 In addition, WES has revealed that ~8% of patients without a definitive causative mutation harbor novel candidate mutations.5 Moreover, for some cases of undiagnosed rare diseases, a new entity has been established among patients whose disorder was previously indistinguishable from other diseases expressing similar phenotypes. Recent studies reported patients with de novo KAT6A mutations among individuals diagnosed with known rare autosomal dominant diseases.6–8 Although those patients had similar phenotypes, the features were nonspecific. Thus, it is difficult to distinguish this disease from other diseases based on clinical findings alone.
In this report, we describe three siblings with ID or global developmental delay with a KAT6A heterozygous nonsense mutation that was potentially transmitted from one of their parents as a germline mosaicism. This result reinforces ‘germline mosaicism’ in genetic counseling for patients with de novo mutation.
Materials and methods
Subjects
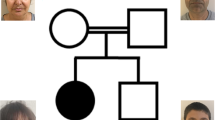
Three Japanese brothers from nonconsanguineous parents were analyzed (Figure 1). The brothers exhibited ID and several common features, suggesting that the underlying cause was genetic. The three siblings, their unaffected brother, and their parents were subjected to genetic testing as part of the Initiative on Rare Undiagnosed Diseases in Pediatrics project in Japan. This study protocol has been approved by the Committee for Ethical Issues on Human Genome and Gene Analysis at Nagasaki University. All genetic analyses were performed in the Department of Human Genetics at Nagasaki University.
Whole-exome sequencing
Peripheral blood was obtained with written informed consent and DNA was extracted using the QIAamp DNA Maxi kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Four subjects, the parents (II-1 and II-7), an affected child (III-1), and an unaffected child (III-2), underwent WES to screen for the candidate causative mutation (Figure 1). Coding exons were captured with the SureSelect XT AUTO HUMAN ALL Exon V5 kit (Agilent Technology, Santa Clara, CA, USA) and sequenced on a HiSeq2500 system (Illumina, San Diego, CA, USA) in rapid mode with 101 bp paired-end reads. Reads were aligned to GRCh37/hg19 with Novoalign (Novocraft Technologies, Selangor, Malaysia) and duplicate reads that were excluded from the following analysis were marked with Novosort software (Novocraft Technologies). Local realignment and variant calling were performed by the Genome Analysis Toolkit.9 Generated variant call format files were processed in the following two manners. First, trio-based VCS format files were filtered to extract the de novo, homozygous, and X-linked mutations followed by annotation with ANNOVAR.10 Second, variant call format files were annotated with ANNOVAR then filtered to extract compound heterozygous mutations. This process excluded variants with allele frequencies >0.5% in the Exome Aggregation Consortium (http://exac.broadinstitute.org/), NHLBI GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), Human Genetic Variation Database (http://www.hgvd.genome.med.kyoto-u.ac.jp), or the database of Tohoku Medical Megabank (http://www.dist.megabank.tohoku.ac.jp). Three de novo mutations identified in WES were amplified by PCR in all family members (II-1, II-7, III-1, III-2, III-3, and III-4) followed by direct sequencing to confirm the existence and linkage to phenotype. The primers designed by Primer3Plus (http://www.bioinformatics.nl/primer3plus) were as follows: KAT6A_F, 5′- ATCTCAAACGTGGGTTCTAA-3′, KAT6A_R, 5′- ATGTGCTAATTCTATTTGGT-3′; CEP89_F, 5′- AACTGGGAACATAGAAAACA-3′, CEP89_R, 5′- CAGTGTCAAGTGTTAAGTGA-3′; and CDX4_F, 5′- TCCAATTTCGCTGCGGCACC-3′, CDX4_R, 5′- AGGGCCCAAGTTGCTGTAGTC-3′.
Results
Clinical features
Individual III-1 was a 14-year-old boy who was born at 40 weeks of gestation with a birth weight of 2,914 g. He was intubated for breathing problems soon after birth due to persistent pulmonary hypertension and was hospitalized for 2 months. He sat unassisted at 12 months and walked at 22 months of age. He spoke at the age of 3, but his vocabulary was limited to a few short nursery words. He underwent surgery for undescended testes and inguinal hernia at 10 months of age. At the age of 7, his height was 115.2 cm (−0.63 standard deviation (SD)), his weight was 18.8 kg (−0.89 SD), and his head circumference was 49.6 cm (−1.47 SD). His intelligence quotient was 13 at 13 years of age.
Individual III-3 was a 10-year-old boy who was born at 37 weeks of gestation with a birth weight of 2,910 g. He had meconium aspiration syndrome and stayed in an incubator for 2 weeks. He was repeatedly admitted to hospital with recurrent pulmonary infection during his first year of life. He sat unassisted at 18 months, walked at 30 months old, and spoke at the age of 5. At 2 years of age, his height was 79 cm (−1.8 SD), his weight was 7.63 kg (−3.25 SD), and his head circumference was 45 cm (−1.72 SD). His intelligence quotient was 26 at 10 years of age.
Individual III-4 was a 10-month-old boy who was born at 35 months of gestation with a birth weight of 2,270 g. He had an atrial septal defect and mild laryngomalacia. He cannot sit without assistance.
All three affected siblings have the clinical triad characteristic of the family involving mild to moderate scaphocephaly, micrognathia, and low-set ears. In addition, feeding difficulties and axial hypotonia are also observed. Individuals III-1 and III-3 have strabismus (exotropia) and abnormal teeth (malocclusions). Their blood cell counts and serum levels of liver enzymes, creatinine, and electrolytes are within normal ranges. Computed tomography scanning revealed no evidence of intracranial abnormalities.
WES and Sanger sequencing
The mean depth of coverage for each individual in WES is as follows: II-1, 74×; II-7, 104×; III-1, 74×; III-2, 77×. Trio analysis revealed that three genes, KAT6A, CEP89, and CDX4, have de novo heterozygous single nucleotide variations in the affected individual III-1 but not in unaffected III-2. Sanger sequencing of all family members (II-1, II-7, III-1, III-2, III-3, and III-4) detected the KAT6A mutation in affected boys but not in the unaffected boy or the parents. This de novo nonsense mutation, c.3070C>T (p.R1024*, ENST00000406337; chr8:41795056G>A on hg19) (Figure 2), was previously reported in two patients.7,8 The phenotypes of our cases are similar to those cases, and we concluded that this mutation is causative of disease in the present family (OMIM: 616268) (Table 1).
Discussion
In this report, we describe three siblings with ID or global developmental delay caused by a KAT6A mutation. To date, 17 cases with ID or global developmental delay from 16 families with KAT6A mutations have been reported.6–8 A recent large-scale study including more than 7,000 individuals with neurodevelopmental disorder identified 11 patients with potentially damaging KAT6A de novo mutations.3 This finding may reflect the actual frequency of the disease.
The main symptoms in our cases involved ID and developmental delay with very limited verbal development, which is consistent with previous reports of patients with KAT6A mutations. Craniofacial abnormalities, especially a small head circumference (some cases met the criteria of microcephaly), are noted at a high frequency and are relatively characteristic.6–8 Low-set ears were observed in all our patients and in 5/7 cases reported by Tham et al.6 Feeding difficulties and axial hypotonia were also observed in most patients, and many patients had congenital heart disease and strabismus. Scaphocephaly was common among our patients but has not previously been observed in other reported cases. Teeth abnormalities and micrognathia are common in patients with the KAT6A p.R1024* mutation. Although several common features can be recognized in individuals with KAT6A mutations, these findings are nonspecific, making the disorder indistinguishable from similar diseases with ID.
Analysis of parental genotypes in the present study indicated that the KAT6A mutation of our patients is de novo but that it must have been transmitted by parental germline mosaicism given its consecutive occurrence. Germline mosaicism refers to variation in the genomes of germline cells within an individual.11 Although it can be beneficial to define the mosaic rate of gametes in such cases, we did not do so, because it is practically impossible to use ova for research purposes. Recent deep sequencing or droplet digital PCR techniques are used to detect a very low prevalence of somatic mosaicism in the blood;12,13 however, this technology is not useful to determine the germline mosaic rate. Acuna-Hidalgo et al.13 reported that the genome-wide analysis of putative de novo mutations in the proband detected 4/4,081 variants in the blood of one of the parents. In our exome analysis, the mutation was not detected within a read depth of 89 in the father (II-1) and 113 in the mother (II-7).
KAT6A, which is also known as MOZ or MYST3 on chromosome 8q11.21, encodes a member of the MYST family of histone acetyl-transferases consisting of 2004 amino acids. KAT6A was first described as a fusion protein in patients with acute myeloid leukemia14 and all members of this family have a histone acetyl-transferase domain that acetylates the histone lysine residue and promotes transcription.15 KAT6A is involved in development through the regulation of Hox gene expression.16 In zebrafish, the KAT6A ortholog specifies segmental identity in the pharyngeal arches.17,18 Kastumoto et al.19 described the Kat6a homozygous knockout mouse model, in which exon 2 containing the first ATG of Kat6a was replaced with a neo gene cassette, resulting in embryonic lethality and a lack of hematopoietic stem cells. Another homozygous knockout model, in which the neo coding sequence was inserted into exon 16, was also lethal, exhibiting craniofacial and heart abnormalities with normal hematopoietic cells.16,20 Although no description exists for heterozygous knockout mouse model defects, the craniofacial and heart abnormalities commonly identified in patients with KAT6A could correspond to this model, except for hematopoietic cell aberrations.
In conclusion, we report three siblings with a KAT6A mutation and ID. This is the first report of KAT6A mutations in an Asian population, and this report also describes the first case of germline mosaicism of the disease. To date, this disease can only be diagnosed after genome-wide testing and the observation of particular craniofacial characteristics. However, the widespread use of WES will lead to an accumulation of patient clinical data that could provide useful information about the diagnosis, prognosis, and future strategies for treating the disease.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S . Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil 2011; 32: 419–436.
Kaufman L, Ayub M, Vincent JB . The genetic basis of non-syndromic intellectual disability: a review. J Neurodev Disord 2010; 2: 182–209.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–438.
Yang Y, Muzny DM, Xia F, Niu Z, Person R, Veeraraghavan N et al. molecular findings among patients referred for clinical whole- exome sequencing yaping. JAMA 2014; 312: 1870–1879.
Farwell Hagman KD, Shinde DN, Mroske C, Smith E, Radtke K, Shahmirzadi L et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med 2017; 19: 224–235.
Tham E, Lindstrand A, Santani A, Malmgren H, Nesbitt A, Dubbs HA et al. Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am J Hum Genet 2015; 96: 507–513.
Arboleda VA, Lee H, Dorrani N, Zadeh N, Willis M, Macmurdo CF et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet 2015; 96: 498–506.
Millan F, Cho MT, Retterer K, Monaghan KG, Bai R, Vitazka P et al. Whole exome sequencing reveals de novo pathogenic variants in KAT6A as a cause of a neurodevelopmental disorder. Am J Med Genet A 2016; 170: 1791–1798.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Gajecka M . Unrevealed mosaicism in the next-generation sequencing era. Mol Genet Genomics 2015; 291: 513–530.
Nakashima M, Takano K, Tsuyusaki Y, Yoshitomi S, Shimono M, Aoki Y et al. WDR45 mutations in three male patients with West syndrome. J Hum Genet 2016; 61: 653–661.
Acuna-Hidalgo R, Bo T, Kwint MP, Van De Vorst M, Pinelli M, Veltman JA et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet 2015; 97: 67–74.
Borrow J, Stanton VP, Andresen JM, Becher R, Behm FG, Chaganti RS et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996; 14: 33–41.
Champagne N, Pelletier N, Yang XJ . The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene 2001; 20: 404–409.
Voss AK, Collin C, Dixon MP, Thomas T . Moz and retinoic acid coordinately regulate H3K9 acetylation, hox gene expression, and segment identity. Dev Cell 2009; 17: 674–686.
Miller CT, Maves L, Kimmel CB . moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development 2004; 131: 2443–2461.
Kong Y, Grimaldi M, Curtin E, Dougherty M, Kaufman C, White RM et al. Neural crest development and craniofacial morphogenesis is coordinated by nitric oxide and histone acetylation. Chem Biol 2014; 21: 488–501.
Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev 2006; 20: 1321–1330.
Voss AK, Vanyai HK, Collin C, Dixon MP, McLennan TJ, Sheikh BN et al. MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev Cell 2012; 23: 652–663.
Acknowledgements
We thank the patients and their family for participating in the study. We also thank Chisa Hayashida for technical support with next generation sequencing. This work was supported by a part of the Initiative on Rare Undiagnosed Diseases in Pediatrics project (KY) and Practical Research Project for Rare/Intractable Diseases from the Agency for Medical Research and Development; Grant-in-Aid for Scientific Research (B) (KY) and (C) (AK) from the Japan Society for the Promotion of Science; Takeda Scientific Foundation (KY).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Satoh, C., Maekawa, R., Kinoshita, A. et al. Three brothers with a nonsense mutation in KAT6A caused by parental germline mosaicism. Hum Genome Var 4, 17045 (2017). https://doi.org/10.1038/hgv.2017.45
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2017.45
This article is cited by
-
Five new cases of syndromic intellectual disability due to KAT6A mutations: widening the molecular and clinical spectrum
Orphanet Journal of Rare Diseases (2020)
-
KAT6A Syndrome: genotype–phenotype correlation in 76 patients with pathogenic KAT6A variants
Genetics in Medicine (2019)