Abstract
Waardenburg syndrome (WS) is a dominantly inherited, genetically heterogeneous auditory-pigmentary syndrome characterized by non-progressive sensorineural hearing loss and iris discoloration. By whole-exome sequencing (WES), we identified a nonsense mutation (c.598C>T) in PAX3 gene, predicted to be disease causing by in silico analysis. This is the first report of genetically diagnosed case of WS PAX3 c.598C>T nonsense mutation in Chinese ethnic origin by WES and in silico functional prediction methods.
Similar content being viewed by others
Waardenburg syndrome (WS) is a rare autosomal dominant disorder with an estimated prevalence of 1:42,000.1 It is characterized by congenital sensorineural hearing loss and depigmentation of the eyes, hair and skin, deafness, with or without dystopia canthorum (a lateral displacement of the inner canthi of the eyes),2,3,4 grouped into four subtypes (WS1–4). First reported in Europe,5 this disease has also been commonly reported in other ethnic groups,6,7 but it is rarely been reported in Chinese population.8
WS rarity and genetic heterogeneity limited the ability to make accurate clinical diagnosis in individual patients. Presently, seven genes are related to WS: PAX3 (encoding the paired box 3 transcription factor), MITF (microphthalmia-associated transcription factor), EDNRB (endothelin receptor type B), EDN3 (endothelin 3), SNAI2 (snail homolog 2), SOX10, and recently reported KITLG (encoding KIT ligand).9 As a result, the accurate diagnosis of WS is difficult to achieve by traditional genetic testing methods, for example, Sanger sequencing. In this paper, a Chinese family affected by WS1 is reported, whole-exome sequencing (WES) was performed and a nonsense mutation in PAX3 gene in Chinese population was discovered.
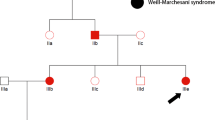
In the present study, three affected patients (I-2, II-2, III-1) and one unaffected member (I-1) from an isolated village in Inner Mongolia in China were involved in our study (Figure 1a). The proband (I-2) was found when she sought treatment due to pneumonia. Dystopia canthorum was present in all patients, but the symptom of III-1 was less severe. Pigmentary disturbance of iris was present in two patients (I-2, II-2) (Figure 1b). Hair hypopigmentation was present in two patients (I-2, II-2), while only one patient (II-2) had sensorineural hearing loss. The clinical features of the proband suggest WS; however, other disorders associated with hearing loss cannot be excluded.
Whole-exome sequencing was performed in order to identify the causal genes. DNA was isolated from peripheral blood by standard procedures (Tiangen, China. Cat. No. DP348). Whole-exome enrichment was performed using SureSelect XT Target Enrichment System (51 Mb) according to the manufacturer’s protocols (Agilent, Santa Clara, CA, USA). Captured libraries were loaded onto the HiSeq 2500 platform (Illumina, San Diego, CA, USA). Workflow recommended by the Genome Analysis Tool kit (GATK) was applied to analyze the variants. Reads were aligned to the human reference genome (UCSC GRCh37/hg19) by using the Burrows-Wheeler Aligner (BWA v0.7.12).10 Presumed PCR duplicates were removed using Picard’s MarkDuplicates. Local alignment optimization and base quality recalibration was performed with the GATK. Indels and single-nucleotide variants were identified by means of the GATK HaplotypeCaller and Unified Genotyper algorithms. Variants were functionally annotated and filtered using our cloud-based rare disease NGS analysis platform (https://www.gene.ac/) with build in public databases (dbSNP, OMIM, ESP, Clinvar, 1,000 Genomes,11 and ExAC)12,13 and HGMD Professional database. Exonic sequence alterations and intronic variants at exon-intron boundaries, with unknown frequency or minor allele frequency (MAF) <1% and not present in the homozygous state in those databases were retained.
A mean depth of target region of 121.81× was achieved, and 86% of targeted bases were read >30 times by whole-exome sequencing. A mean of only 7.11% bases in the targeted coding regions had <10× coverage, and a mean of only 5.08% bases had <4× coverage. A total of 584,454 SNPs were identified (see Supplementary Dataset). We identified mutation c.598C>T in exon5 of PAX3 gene was likely to be pathogenic, which is a nonsense mutation causing the glutamine to stop codon substitution at codon 200 (Figure 2). This variant of Irish origin was firstly reported in 1995 by Baldwin et al.,2,14 and documented in LOVD database (http://grenada.lumc.nl/LOVD2/WS/). It was predicted to be disease-causing by in silico analysis using Mutation Taster (http://www.mutationtaster.org/), with P-value of 0.99969, suggesting a high possibility of protein functional alteration, which has not been described in previous report. This variant co-segregates with all affected individuals in the family and was not observed in unaffected family members. Gln200 is localized in a phylogenetically conserved HD domain and is evolutionarily conserved from mouse to humans (Figures 2b and c). This mutation was further validated by Sanger sequencing (Figure 2a). The amplicon was directly sequenced using the ABI BigDye Terminator Sequencing Kit (Applied Biosystems, Shanghai, China) and an automated capillary sequencer (ABI 3730×l; Applied Biosystems). Sequence electropherograms were analyzed using the Chromas software.
(a) Sequence chromatograms of the family: a heterozygous change, c.598C>T, was identified in patients I-2, II-2 and III-1) (b) Schematic diagram of the PAX3 gene. The boxes indicate the structure of the PAX3 protein. The arrow of patient indicates the position of mutation in PAX3 protein. (c) Cross-species multiple alignment of PAX3 protein sequences, showing the evolutionally conserved sequence around position Q200.
Recently, WES has been demonstrated as a powerful approach to elucidate the genetic basis of rare Mendelian disorders with unknown etiology.15,16,17,18 In addition, WES is increasingly being utilized as a diagnostic tool for specific genetic diseases with significant genetic and phenotypic heterogeneity.15 The progression of genomic technologies over the past few years has led to a disruptive shift in our approach to the identification of new Mendelian disorders for numerous previously unresolved rare disorders.18,19 WES is appropriate because of lower turnaround time, significantly reduced cost, and more focused and comprehensive coverage of target genes, and easier data analysis compared with whole genome sequencing.
In conclusion, we identified a nonsense pathogenic variant (c.598C>T; p.Gln200*) in the PAX3 gene that has not been reported in Chinese population before. This report will contribute to a better WS mutation spectrum identified so far. We also demonstrated that WES could be a powerful approach to identify pathogenic variants in patients with WS.
References
References
Read AP, Newton VE . Waardenburg syndrome. J Med Genet 1997; 34: 656–665.
Baldwin CT, Hoth CF, Macina RA, Milunsky A . Mutations in PAX3 that cause Waardenburg syndrome type I: ten new mutations and review of the literature. Am J Med Genet 1995; 58: 115–122.
Eigelshoven S, Kameda G, Kortum AK, Hubsch S, Angerstein W, Singh P et al. Waardenburg syndrome type I with heterochromia iridis and circumscribed hypopigmentation of the skin. Pediatr Dermatol 2009; 26: 759–761.
Pardono E, van Bever Y, van den Ende J, Havrenne PC, Iughetti P, Maestrelli SR et al. Waardenburg syndrome: clinical differentiation between types I and II. Am J Med Genet A 2003; 117A: 223–235.
Waardenburg PJ . A new syndrome combining developmental anomalies of the eyelids, eyebrows and nose root with pigmentary defects of the iris and head hair and with congenital deafness. Am J Hum Genet 1951; 3: 195–253.
Choi JH, Moon SK, Lee KH, Lew HM, Chang YH . Three cases of Waardenburg syndrome type 2 in a Korean family. Korean journal of ophthalmology. KJO 2004; 18: 185–189.
Jang MA, Lee T, Lee J, Cho EH, Ki CS . Identification of a novel de novo variant in the PAX3 gene in waardenburg syndrome by diagnostic exome sequencing: the first molecular diagnosis in Korea. Ann Lab Med 2015; 35: 362–365.
Sun L, Li X, Shi J, Pang X, Hu Y, Wang X et al. Molecular etiology and genotype-phenotype correlation of Chinese Han deaf patients with type I and type II Waardenburg Syndrome. Sci Rep 2016; 6: 35498.
Zazo Seco C, Serrao de Castro L, van Nierop JW, Morin M, Jhangiani S, Verver EJ et al. Allelic mutations of KITLG, encoding kit ligand, cause asymmetric and unilateral hearing loss and waardenburg syndrome type 2. Am J Hum Genet 2015; 97: 647–660.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Bahcall OG . Genetic variation: ExAC boosts clinical variant interpretation in rare diseases. Nature reviews Genetics 2016; 17: 584.
Song W, Gardner SA, Hovhannisyan H, Natalizio A, Weymouth KS, Chen W et al. Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet Med 2016; 18: 850–854.
Farrer LA, Grundfast KM, Amos J, Arnos KS, Asher JH Jr, Beighton P et al. Waardenburg syndrome (WS) type I is caused by defects at multiple loci, one of which is near ALPP on Chromosome 2:First report of the WS consortium. Am J Hum Genet 1992; 50: 902–913.
Ku CS, Cooper DN, Polychronakos C, Naidoo N, Wu M, Soong R . Exome sequencing: dual role as a discovery and diagnostic tool. Ann Neurol 2012; 71: 5–14.
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014; 312: 1880–1887.
Lohmann K, Klein C . Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics 2014; 11: 699–707.
Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010; 42: 30–35.
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009; 461: 272–276.
Data Citations
Xiao, Han HGV Database (2017) http://dx.doi.org/10.6084/m9.figshare.hgv.1370
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information for this article can be found on the Human Genome Variation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, D., Zhao, N., Wang, J. et al. Whole-exome sequencing analysis of Waardenburg syndrome in a Chinese family. Hum Genome Var 4, 17027 (2017). https://doi.org/10.1038/hgv.2017.27
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2017.27
This article is cited by
-
A novel de novo mutation in COL2A1 leading to spondyloepiphyseal dysplasia congenita in a Chinese family
Human Genome Variation (2018)