Abstract
Stickler syndrome (STL) is an autosomal, dominantly inherited, clinically variable and genetically heterogeneous connective tissue disorder characterized by ocular, auditory, orofacial and skeletal abnormalities. We conducted targeted resequencing using a next-generation sequencer for molecular diagnosis of a 2-year-old girl who was clinically suspected of having STL with Pierre Robin sequence. We detected a novel heterozygous missense mutation, NM_001854.3:n.4838G>A [NM_001854.3 (COL11A1_v001):c.4520G>A], in COL11A1, resulting in a Gly to Asp substitution at position 1507 [NM_001854.3(COL11A1_i001)] within one of the collagen-like domains of the triple helical region. The same mutation was detected in her 4-year-old brother with cleft palate and high-frequency sensorineural hearing loss.
Similar content being viewed by others
STL is a clinically and genetically heterogeneous group of collagenopathies characterized by ocular, auditory, skeletal and orofacial abnormalities.1 At least three subtypes of an autosomal dominant form of STL have been delineated: types 1 (STL 1, OMIM 108300), 2 (STL 2, OMIM 604841) and 3 (STL 3, OMIM 184840); these subtypes are a result of heterozygous mutations in COL2A1 (OMIM 120140), coding type II fibrillar collagen, COL11A1 (OMIM 120280), coding the α1 chain of type XI fibrillar collagen and COL11A2 (OMIM 120290), coding the α2 chain of type XI fibrillar collagen, respectively.2–4 Mutations in other genes, including biallelic mutations in COL9A1, COL9A2 and COL9A3 for an autosomal recessive form, are also known to be associated with this disease.1 Variable phenotypic expression of STL occurs both within and among families.1,5
The diagnosis of STL is clinically based, but no consensus has been reached on the minimal clinical diagnostic criteria.1 As differential diagnosis is sometimes difficult owing to phenotypic variability, genetic testing facilitates diagnosis in some cases. Serial single-gene molecular genetic testing using standard Sanger sequencing based on an individual’s clinical findings and family history is routinely used to identify pathogenic mutations in clinical testing. However, the limitations of this genetic testing strategy, including low throughput and high cost, make the use of multigene panels laborious, time consuming and expensive. These limitations are particularly prominent when a target gene contains multiple exons (e.g., collagen genes), multiple genes are responsible for one disease due to genetic heterogeneity (e.g., STL) or multiple diseases with overlapping features caused by different genes exist.6 Disease-related gene panels or targeted-exome sequencing (TES) of candidate and/or known pathogenic genes using next-generation sequencing (NGS) technology can change the landscape of medical sequencing to diagnose patients with congenital disorders such as STL.7,8 We report a novel heterozygous and possibly pathogenic variation in COL11A1, NM_001854.3:n.4838G>A [NM_001854.3 (COL11A1_v001):c.4520G>A], detected by targeted-NGS in our study of a Japanese patient with STL using a targeted-exome panel comprising the coding regions of 4,813 clinical phenotype-associated genes, including STL-related genes.
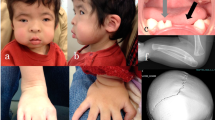
The proband (IV:2, Figure 1) was a 2-year-old girl of healthy, non-consanguineous parents who was born after 40 weeks of gestation. At birth, her weight, height and occipitofrontal circumference (OFC) were 3,002 g (+0 s.d), 50 cm (+0.8 s.d) and 32.5 cm (−0.3 s.d), respectively. She was born with a cleft palate, and automated auditory brainstem response (AABR) testing suggested bilateral hearing impairment. At 2 months of age, she visited the Tokyo Medical and Dental University Hospital for cleft palate treatment. At this time, Pierre Robin sequence (U-shaped cleft palate, micro/retrognathia and glossoptosis) and her brother’s cleft palate were indicated. At 6 months of age, she was clinically diagnosed with STL according to her craniofacial anomalies as well as her older brother’s clinical characteristics associated with this disease, as described below. Plastic surgery for cleft palate was successfully performed. Eardrum tube was placed to alleviate otitis media with effusion. Her motor and mental development was normal. Bilateral mild high-frequency sensorineural hearing loss (HFSNHL) was revealed by audiometric testing, but no abnormality was revealed on ophthalmological evaluation up to 2 years of age.
Her older brother (IV:1, Figure 1) was a 4-year-old boy who was born after 40 weeks of gestation. At birth, his weight, height and OFC were 3,060 g (+0.1 s.d), 48 cm (−0.5 s.d) and 35 cm (+1.2 s.d), respectively. He was born with a cleft palate, and AABR testing suggested bilateral hearing impairment. At 1 year of age, plastic surgery for cleft palate was successfully performed. Eardrum tube was placed to alleviate otitis media with effusion. His motor and mental development was normal. Owing to his younger sister’s cleft palate, physical and radiographic examinations were performed at 34 months of age. A flat midface with nasal bridge hypoplasia was indicated at that age. Radiographs taken at that age revealed metaphyseal widening of the femoral neck and mild platyspondyly. Although the otitis media improved, audiometric testing at that age revealed bilateral HFSNHL. No ophthalmological abnormalities had been observed up to 4 years of age. His clinical characteristics pertaining to orofacial, auditory and skeletal abnormalities were compatible with those of STL. Detailed information on the orofacial, ocular, auditory and skeletal characteristics of the patients’ parents was not available (Figure 1).
The ethics committees of the Tokyo Medical and Dental University and Tokushima University approved this study. After obtaining written consent from the mother of the patients, molecular diagnosis was performed using genomic DNA extracted from the patients’ whole blood. We first performed TES using a TruSight One Sequencing Panel (Illumina, San Diego, CA, USA) and an MiSeq benchtop sequencer (Illumina) for the proband (IV:2). The alignments of sequencing reads to the human reference genome (hg19), duplicate read removal, local realignment around indels, base quality score recalibration, variant calling and annotation were performed as described elsewhere.9 To identify single-nucleotide variations (SNVs), we excluded minor sequence variants with low-allele frequencies, i.e., >0.05 included in the 1000 Genomes Project database (http://www.1000genomes.org/), NHLBI GO Exome Sequencing Project (ESP6500, http://evs.gs.washington.edu/EVS/), Human Genetic Variation Database (HGVD, http://www.genome.med.kyoto-u.ac.jp/SnpDB/) and integrative Japanese Genome Variation Database (iJGVD, https://ijgvd.megabank.tohoku.ac.jp/).10 The detection of copy-number variations (CNVs) using NGS data with a resolution of a single exon to several exons depending on the size of the exons was performed as described elsewhere.11
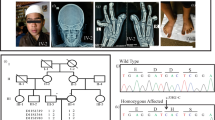
These analyses identified one heterozygous mutation, NM_001854.3:n.4838G>A [NM_001854.3 (COL11A1_v001):c.4520G>A], in exon 61 of COL11A1 (NM_001854.3), resulting in a Gly to Asp substitution at position 1507 [NM_001854.3(COL11A1_i001)] within the eighth collagen-like domain of the triple helical region. This mutation was confirmed by Sanger sequencing, and the same mutation was also found in her older brother (Figure 2a). Primer sequences are available upon request. No other potentially pathogenic mutations, including SNVs and CNVs, were detected in genes potentially responsible for STL or related diseases. This mutation is neither present in human genome variation databases, such as dbSNP138, 1000 Genomes Project, ESP6500, HGVD and iJGVD nor in disease-causing mutation databases such as the Human Gene Mutation Database Professional 2015.4 (HGMD, http://www.hgmd.org/) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). Through in silico analysis using LJB23 databases for non-synonymous variant annotation in ANNOVAR (http://annovar.openbioinformatics.org/en/latest/), NM_001854.3(COL11A1_v001):c.4520G>A is predicted to damage protein function, i.e., SIFT score=0, Polyphen2 score=1 and MutationTaster score=1. The presence of Gly at every third residue (Gly-X-Y) is considered to be essential for the formation of the characteristic collagen triple helix (Figure 2b), and Asp is the most destabilizing residue in the Gly position of the Gly-X-Y repeating pattern in type I collagen, one of the fibrillar collagens.12 In HGMD, single-base substitutions in Gly codons in the triple-helix-encoding regions were observed in 15 among 22 missense mutations of COL11A1 (68.2%). In addition, a Gly to Asp substitution was reported for 2 of those 15 mutations,13,14 suggesting that this is a novel mutation responsible for STL. As parental DNA was not available, it could not be determined whether the mutation had been inherited from one of the parents or developed de novo.
(a) Electropherogram of COL11A1 (NM_001854.3) exon 61 and flanking intron 60 in the affected siblings (IV:1 and IV:2). Red arrowheads denote the mutated base. The DNA and corresponding amino acid sequences of wild-type and mutant COL11A1 alleles are shown. (b) Multiple alignment of the COL11A1 amino acid sequences around codon 1507. The red arrowhead denotes the mutated amino acid. Dots indicate conserved Gly in the Gly-X-Y repeating pattern. Amino acids not matched with those in Homo sapiens are light gray.
As accurate radiographic and clinical examinations, including formal testing of hearing and vision,5 and genetic testing of the parents were not performed, it remains unclear whether the same COL11A1 mutation observed in the siblings had been inherited or produced de novo.1 Although the penetrance of STL is known to be high, variable clinical expression of STL complicates the genetic counseling scenario.5 As it was reported that the COL2A1 frameshift mutation leading to a premature stop codon was detected in family members who were considered healthy in the STL family, mutations in collagen genes may not necessarily result in the full STL phenotype, but may cause a mild phenotype or remain below the clinical expression level.15 At present, it cannot be ruled out that, among the carriers of mutations, subjects may frequently have an atypical phenotype rather than the typically known phenotype of STL.15 Although instances of germline mosaicism have been reported only in STL with COL2A1 mutation, this also remains a possibility.16 Such insights would be helpful for counseling and parental decision making.
References
References
Robin NH, Moran RT, Ala-Kokko L . Stickler syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH et al. (eds). GeneReviews [Internet]. University of Washington: Seattle, WA, USA, 2009 [updated 26 Nov 2014] .
Ahmad NN, Ala-Kokko L, Knowlton RG, Jimenez SA, Weaver EJ, Maguire JI et al. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy). Proc Natl Acad Sci USA 1991; 88: 6624–6627.
Vikkula M, Mariman ECM, Lui VCH, Zhidkova NI, Tiller GE, Goldring MB et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell 1995; 80: 431–437.
Richards AJ, Yates JRW, Williams R, Payne SJ, Pope FM, Scott JD et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in α1(XI) collagen. Hum Mol Genet 1996; 5: 1339–1343.
Snead MP, Yates JRW . Clinical and molecular genetics of Stickler syndrome. J Med Genet 36: 353–359.
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15: 733–747.
Acke FR, Malfait F, Vanakker OM, Steyaert W, De Leeneer K, Mortier G et al. Novel pathogenic COL11A1/COL11A2 variants in Stickler syndrome detected by targeted NGS and exome sequencing. Mol Genet Metab 2014; 113: 230–235.
Kohmoto T, Naruto T, Kobayashi H, Watanabe M, Okamoto N, Masuda K et al. A novel COL11A1 mutation affecting splicing in a patient with Stickler syndrome. Hum Genome Variation 2015; 2: 15043.
Okamoto N, Naruto T, Kohmoto T, Komori T, Imoto I . A novel PTCH1 mutation in a patient with Gorlin syndrome. Hum Genome Variation 2014; 1: 14022.
Naruto T, Okamoto N, Masuda K, Endo T, Hatsukawa Y, Kohmoto T et al. Deep intronic GPR143 mutation in a Japanese family with ocular albinism. Sci Rep 2015; 5: 11334.
Yamaguchi-Kabata Y, Nariai N, Kawai Y, Sato Y, Kojima K, Tateno M et al. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Hum Genome Variation 2015; 2: 15050.
Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JAM, Brodsky B . Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc Natl Acad Sci USA 2000; 97: 4273–4278.
Richards AJ, McNinch A, Martin H, Oakhill K, Rai H, Waller S et al. Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum Mutat 2010; 31: E1461–E1471.
Majava M, Hoornaert KP, Bartholdi D, Bouma MC, Bouman K, Carrera M et al. A report on 10 new patients with heterozygous mutations in the COL11A1 gene and a review of genotype–phenotype correlations in type XI collagenopathies. Am J Med Genet A 2007; 143A: 258–264.
Faber J, Winterpacht A, Zabel B, Gnoinski W, Schinzel A, Steinmann B et al. Clinical variability of Stickler syndrome with a COL2A1 haploinsufficiency mutation: implications for genetic counselling. J Med Genet 2000; 37: 318–320.
Stevenson DA, Vanzo R, Damjanovich K, Hanson H, Muntz H, Hoffman RO et al. Mosaicism in Stickler syndrome. Eur J Med Genet 2012; 55: 418–422.
Data Citations
Imoto, I HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.780
Acknowledgements
Authors would like to thank the patient and her family for participating in this study. This work was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. Also, this work was supported by JSPS KAKENHI Grant Numbers 26293304 (I.I.) and 15K19620 (T.N.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Kohmoto, T., Tsuji, A., Morita, Ki. et al. A novel COL11A1 missense mutation in siblings with non-ocular Stickler syndrome. Hum Genome Var 3, 16003 (2016). https://doi.org/10.1038/hgv.2016.3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2016.3