Abstract
Propionic acidemia (PA) is an inborn error of metabolism, caused by mutations in either the PCCA or PCCB gene, leading to mitochondrial accumulation of propionyl-CoA and its by-products. Here we report a 6-year-old Thai boy with PA who was born to consanguineous parents. Exome sequencing identified a novel homozygous frameshift insertion (c.379_380insA; p.T127NfsX160) in the PCCB gene, expanding its mutational spectrum.
Similar content being viewed by others
Propionic acidemia (PA) is an autosomal recessive metabolic disorder caused by defects in the propionyl-CoA carboxylase (PCC) enzyme. PCC is involved in catabolism of branched-chain amino acids, odd chain fatty acids and cholesterol.1,2 Disrupted function of PCC leads to mitochondrial accumulation of propionyl-CoA and its by-products. A characteristic urine organic acid profile shows 3-hydroxypropionate, methylcitrate, tiglylglycine and propionylglycine. The clinical signs and symptoms include acute deterioration, hypotonia, metabolic acidosis, ketonuria, hypoglycemia and hyperammonemia, which usually present during infancy and early childhood.3 Without treatment, these can progress to coma and death. Patients who survive the neonatal period mostly have episodes of metabolic acidosis and hyperammonemia.4 It has been shown that mutations in either the PCCA (chromosome 13q32) or PCCB (chromosome 3q21-q22) genes encoding alpha or beta subunits of the PCC enzyme, respectively, can cause propionic acidemia.5,6 Mutations in PCCA are heterogeneous without a predominant mutation in the population studied, whereas PCCB has a limited number of mutations in various ethnic groups.5,7 Missense mutations are the most common defects identified in the PCCB gene.7,8 The incidence of PA ranges from 1:50,000 to 1:150,000.9–11 A high frequency of PA has been reported in some populations, for example, 1 in 1,000 among the Greenlandic Inuit population due to a founder effect12 and 1:2,000–5,000 live births in Saudi Arabia partly owing to a relatively high rate of consanguinity.13 To date, at least 81 mutations and 17 polymorphisms have been identified in the PCCA gene, and 86 mutations and 7 polymorphisms in the PCCB gene (http://cbs.lf1.cuni.cz/pcc/pccmain.htm, accessed on January 2015). PCCA mutations have been identified in a small number of Thai patients whereas PCCB mutations have never been reported in Thailand.14
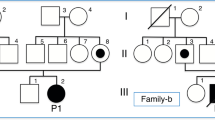
Here we describe a 6-year-old Thai boy with PA who was born to consanguineous parents. The first three children of this family died in infancy owing to metabolic acidosis, but without a definite diagnosis. The fourth one was healthy. The proband was the fifth child of the family. His birth weight was 3.5 kg. Newborn screening using tandem mass spectrometry (MS/MS) showed elevated three-carbon acylcarnitine (C3). Urine organic acid analysis using gas chromatography and mass spectrometry revealed increased urinary excretion of glycine, 3-hydroxypropionate, propionylglycine, tiglyglycine and methylcitrate, a characteristic profile of PA.15 Special formula, l-carnitine and biotin were started since the first week of life. His weight and height continued to be within normal ranges, up to his last visit at the age of 6 years. However, he had delayed development; he started to walk and talk at the age of 4 years.
In this study, we aim to identify the disease-causing mutation of PA in this proband using whole-exome sequencing (WES). Blood samples were obtained. After informed consent, the patient’s genomic DNA was extracted from peripheral blood leukocytes and sent to Macrogen (Seoul, Korea). Four micrograms of DNA sample was enriched by TruSeq Exome Enrichment kit and was sequenced onto Hiseq 2000 (Illumina, San Diego, CA, USA). The raw data per exome were mapped to the human reference genome hg19 using CASAVA v1.7 (Illumina). Variant calling was performed using SAM tools. Total yield of 10,475,843,220 bp was achieved. The capture efficiency varied across the target with 90.5% more than 10X and the mean read depth of target regions was 49×. The sequencing data were mapped to NCBI37 reference human genome (the version used for 1000 Genomes project), NHLBI Exome Variant Server, and in House database. WES of the proband revealed a novel homozygous frameshift insertion (NM_000532: c.379_380insA; p.T127NfsX160) in exon 4 of the PCCB gene.
The variant of interest was validated with Sanger sequencing. PCCBex4-F: TCT ATT TCT TCC TCC CAC TG and PCCBex4-R: GTA CCT AGC TCA GTG TCT TG were used. PCR products were treated with ExoSAP-IT (USP Corporation, Cleveland, OH, USA) and sent for direct sequencing at Macrogen. Sequence data were analyzed using Sequencher (V.5.0; Gene Codes Corporation, Ann Arbor, MI, USA). Sanger sequencing was used to confirm the presence of the identified variant in the patient and his parents. Both parents were heterozygous for the insertion (Figure 1).
Using WES, we successfully identified a mutation in the PCCB gene which has never previously been reported in a patient with PA. WES, next-generation sequencing (NGS), starts with breaking DNA into millions of short fragments and is followed by massively parallel sequencing of all exons in all genes, giving an output of >10 GB in one run. It is suitable for detection of single-nucleotide substitutions, insertions or deletions smaller than 8–10 nucleotides.16 Compared with a conventional mutation detection technique involving PCR–Sanger sequencing, WES is both less time consuming and less expensive to identify mutations in these two genes causing PA. Moreover, it provides an opportunity to find new causative genes and actionable pathogenic variants.17
PCCA has 24 coding exons ranging from 37 to 335 bp in length, whereas PCCB have 15 exons of 57–183 bp.18 The approximate cost of PCR–Sanger sequencing is USD 45 for one exon. Setting up a molecular diagnostic test for PA using conventional sequencing of all these 39 exons would cost almost USD 2,000 in Thailand. The cost of whole human genome sequencing markedly decreases since 2007 from hundreds of millions of dollars to less than ten thousands by NGS.19 For this patient, the cost of WES was less than USD 1,000 and validation of WES with Sanger sequencing was about USD 150. Regarding turnaround time, PCR–Sanger sequencing of 39 exons would take several months, while WES takes only a few weeks.
WES reveals a homozygous frameshift mutation in PCCB expected to result in unstable mRNA, which might be degraded by nonsense-mediated mRNA decay. It has never been previously described. His parents were heterozygous for the mutation. It is absent in 100 exome Thai controls.
In conclusion, we identified a novel homozygous frameshift insertion, c.379_380insA; p.T127NfsX160, in the PCCB gene associated with PA in a Thai family.
References
References
Fenton WA, Gravel RA, Rosenblatt DS . Disorders of propionate and methylmalonate metabolism. In: Scriver CR, Beaudet AR, Sly W, Valle D (eds). The Metabolic and Molecular Bases of Inherited Disease. 8th edn McGraw-Hill: New York, USA, 2001; 2165–2193.
Pena L, Franks J, Chapman KA, Gropman A, Ah Mew N, Chakrapani A et al. Natural history of propionic acidemia. Mol Genet Metab 2012; 105: 5–9.
Dionisi-Vici C, Deodato F, Röschinger W, Rhead W, Wilcken B . ‘Classical’ organic acidurias, propionic aciduria, methylmalonic aciduria and isovaleric aciduria: Long-term outcome and effects of expanded newborn screening using tandem mass spectrometry. J Inherit Metab Dis 2006; 29: 383–389.
Lücke T, Pérez-Cerdá C, Baumgartner M, Fowler B, Sander S, Sasse M et al. Propionic acidemia: unusual course with late onset and fatal outcome. Metabolism 2004; 53: 809–810.
Ugarte M, Pérez-Cerdá C, Rodríguez-Pombo P, Desviat LR, Pérez B, Richard E et al. Overview of mutations in the PCCA and PCCB genes causing propionic acidemia. Hum Mut 1999; 14: 275–282.
Lamhonwah AM, Barankiewicz TJ, Willard HF, Mahuran DJ, Quan F, Gravel RA . Isolation of cDNA clones coding for the alpha and beta chains of human propionyl-CoA carboxylase: chromosomal assignments and DNA polymorphisms associated with PCCA and PCCB genes. Proc Natl Acad Sci USA 1986; 83: 4864–4868.
Rodríguez-Pombo P, Hoenicka J, Muro S, Pérez B, Pérez-Cerdá C, Richard E et al. Human propionyl-coa carboxylase β subunit gene: exon-intron definition and mutation spectrum in Spanish and Latin American propionic acidemia patients. Am J Hum Genet 1998; 63: 360–369.
Desviat LR, Pérez B, Pérez-Cerdá C, Rodríguez-Pombo P, Clavero S, Ugarte M . Propionic acidemia: mutation update and functional and structural effects of the variant alleles. Mol Genet Metab 2004; 83: 28–37.
Chace DH, DiPerna JC, Kalas TA, Johnson RW, Naylor EW . Rapid Diagnosis of Methylmalonic and Propionic Acidemias: Quantitative Tandem Mass Spectrometric Analysis of Propionylcarnitine in Filter-Paper Blood Specimens Obtained from Newborns. Clin Chem 2001; 47: 2040–2044.
Yorifuji T, Kawai M, Muroi J, Mamada M, Kurokawa K, Shigematsu Y et al. Unexpectedly high prevalence of the mild form of propionic acidemia in Japan: presence of a common mutation and possible clinical implications. Hum Genet 2002; 111: 161–165.
Niu D-M, Chien Y-H, Chiang C-C, Ho H-C, Hwu W-L, Kao S-M et al. Nationwide survey of extended newborn screening by tandem mass spectrometry in Taiwan. J Inherit Metab Dis 2010; 33: 295–305.
Ravn K, Chloupkova M, Christensen E, Brandt NJ, Simonsen H, Kraus JP et al. High incidence of propionic acidemia in greenland is due to a prevalent mutation, 1540insCCC, in the gene for the β-subunit of propionyl CoA carboxylase. Am J Hum Genet 2000; 67: 203–206.
Rashed M, Ozand Pt Fau—al Aqeel A, al Aqeel A Fau—Gascon GG, Gascon GG . Experience of King Faisal Specialist Hospital and Research Center with Saudi organic acid disorders. Brain Dev 1994; 16: Suppl: 1–6.
Vatanavicharn N, Liammongkolkul S, Sakamoto O, Kamolsilp M, Sathienkijkanchai A, Wasant P . Clinical characteristics and mutation analysis of propionic acidemia in Thailand. World J Pediatr 2014; 10: 64–68.
Shotelersuk V, Srivuthana S, Wacharasindhu S, Dhamcharee V, Jaruratanasirikul S, Pangkanon S et al. Establishing gas chromatography-mass spectrometry to diagnose organic acidemias in Thailand. Southeast Asian J Trop Med Public Health 2000; 31: 566–570.
Biesecker LG, Biesecker BB . An approach to pediatric exome and genome sequencing. Cur Curr Opin Pediatr 2014; 26: 639–645.
Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574.
Campeau E, Desviat LR, Leclerc D, Wu X, Pérez B, Ugarte M et al. Structure of the PCCA gene and distribution of mutations causing propionic acidemia. Mol Genet Metab 2001; 74: 238–247.
Collins FS, Hamburg MA . First FDA authorization for next-generation sequencer. N Engl J Med 2013; 369: 2369–2371.
Data Citations
Suphapeetiporn, Kanya HGV Database (2015) http://dx.doi.org/10.6084/m9.figshare.hgv.693
Acknowledgements
We thank Supasak Kulawonganunchai for exome-sequencing data analysis and Somporn Liammongkolkul, Division of Medical Genetics, Department of Pediatrics, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand for performing the first organic acid analysis for diagnosis of this patient. This study was supported by the Ratchadapiseksomphot Endowment Fund (RES560530177-HR, RES560530246-AS, RES560530253-AS, WCU-58-024-AS) and the Faculty Research Grant of the Faculty of Dentistry (DRF56018), Chulalongkorn University, and the Thailand Research Fund (RTA5680003, TRG5780209).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Porntaveetus, T., Srichomthong, C., Suphapeetiporn, K. et al. A novel PCCB mutation in a Thai patient with propionic acidemia identified by exome sequencing. Hum Genome Var 2, 15033 (2015). https://doi.org/10.1038/hgv.2015.33
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2015.33
This article is cited by
-
Identification and characterization of the largest deletion in the PCCA gene causing severe acute early-onset form of propionic acidemia
Molecular Genetics and Genomics (2023)