Abstract
Algeria represents a reservoir of genetic diversity with local sheep breeds adapted to a large range of environments and showing specific features necessary to deal with harsh conditions. This remarkable diversity results from the traditional management of dryland by pastoralists over centuries. Most of these breeds are poorly productive, and the economic pressure leads farmers to realize anarchic cross-breeding (that is, not carried out in the framework of selection plans) with the hope to increase animal’s conformation. In this study, eight of the nine local Algerian sheep breeds (D’men, Hamra, Ouled-Djellal, Rembi, Sidaoun, Tazegzawt, Berber and Barbarine) were investigated for the first time by genome-wide single-nucleotide polymorphism genotyping. At an international scale, Algerian sheep occupied an original position shaped by relations with African and European (particularly Italian) breeds. The strong genetic proximity with Caribbean and Brazilian breeds confirmed that the genetic make-up of these American breeds was largely influenced by the Atlantic slave trade. At a national scale, an alarming genetic dilution of the Berber (a primitive breed) and the Rembi was observed, as a consequence of uncontrolled mating practices with Ouled-Djellal. A similar, though less pronounced, phenomenon was also detected for the Barbarine, another ancestral breed. Genetic originality appeared to be better preserved in Tazegzawt, Hamra, D’men and Sidaoun. These breeds should be given high priority in the establishment of conservation plans to halt their progressive loss. For Berber and Barbarine that also occur in the bordering neighbor countries, urgent concerted transnational actions are needed.
Similar content being viewed by others
Introduction
North African livestock has evolved over centuries managed in a traditional way. Furthermore, in the dryland mountain ecosystems (steppes and highlands) encountered in this area, combinations of a wide range of variables (temperature, rainfall, altitude, topography, geology, ground vegetation, soil nutrients and so on) result in a patchwork of nested habitats. All these conditions have enabled the emergence of highly diverse breeds, adapted to a range of dry environments. According to the Food and Agriculture Organization (FAO et al., 2011), one quarter of the world’s biodiversity hot spots are found in such extremely heterogeneous environment. More broadly, livestock of the Near East and North Africa region is regarded as containing unique breeds in terms of genetic diversity (Iniguez, 2005).
Unfortunately, up to 30% of global mammalian and avian livestock breeds are at risk of being lost (FAO, 2013); taking action to halt the erosion of this genetic heritage is an urgent need (Taberlet et al., 2008). The initial emergency is to determine most critical areas, knowing that in spite of its uniqueness, the livestock of Near East and North Africa region is still largely unknown. Moreover, in a climate change perspective, the genetic make-up of these local breeds, showing adaptive traits of particular importance, should be explored and preserved as a priority.
Algeria is the biggest producer of small ruminants in North Africa, together with Morocco (22 million head in each country). Sheep breeding represents 80% of the total domestic animal production. Among the nine local Algerian sheep breeds (D’men, Hamra, Ouled-Djellal, Rembi, Taâdmit, Sidaoun, Tazegzawt, Berber and Barbarine), Ouled-Djellal, which is considered more productive, is taking the monopole as it accounts for >63% of the Algerian sheep population (MADR, 2010). Crosses between Ouled-Djellal and the other Algerian breeds are a current practice of farmers, who are under increasing pressure to improve productivity (Madani et al., 2003). Many of the local Algerian breeds are under high risk of disappearance (Iniguez, 2005), either because of genetic dilution induced by cross-breeding with the Ouled-Djellal or because they are abandoned and suffer from high census contraction (Madani et al., 2003). A recent study of Gaouar et al. (2015), using microsatellites markers and considering six of the nine local breeds, showed that Rembi and Taâdmit were highly admixed with Ouled-Djellal, whereas other breeds appeared more preserved.
In this study, we assessed the genetic diversity of eight Algerian breeds (only the Taâdmit was not considered here; this breed is represented by <100 individuals in governmental farms, and hence we decided to focus efforts on the remaining breeds) using Illumina OvineSNP50K Genotyping BeadChip. The aims of the study were: (1) to provide a comprehensive view of the genetic diversity of Algerian sheep by furthering the investigations began by Gaouar et al. (2015), using a more powerful tool and a wider breed repertoire, also including three breeds (Berber, Barbarine and Tazegzawt) considered here for the first time from a genetic point of view; and (2) to bring new information to the history of the sheep domestication by exploring the relationships of Algerian sheep in an international context (by comparison with world-wide panel of sheep breeds genotyped by the sheep HapMap Project (Kijas et al., 2012) and Italian breeds provided by the BiOvIta Consortium (Ciani et al., 2014)).
Materials and methods
Breeds and sampling design
We sampled eight Algerian breeds, out of which five (Hamra, Rembi, Ouled-Djellal, D’men and Sidaoun) have been described in detail in Gaouar et al. (2015). Three Algerian local breeds (Tazegzawt, Berber and Barbarine) are investigated here for the first time from a genetic point of view. Indeed, the sampling of these breeds is particularly difficult given the growing shortage of flocks. According to Sagne (1950) and Chellig (1992), Algerian sheep takes its origin from three primitive groups: (1) Berber, (2) Arab and (3) Barbarine. The Berber group is considered the ancestor of the North-African sheep, as illustrated on rock paintings of the Stone Age (Sarson, 1973), and it would be the source of the two current breeds: Berber and Hamra. Berber is considered to be the most ancient and primitive sheep breed of the Maghreb. It is a small rustic breed, suited to poor pasture and reared in the Kabylie Mountains of Algeria. Sagne (1950) reported that Herodotus documents suggested the presence of this breed in Kabylie, 3000 years BC. The Arab group (including Ouled-Djellal and Rembi) was presumably introduced into the country during the Zenete invasions (Sagne, 1950; Turries, 1976) (that is, after the Roman occupation and before the Arab conquest). Alternatively, following Trouette (1933), this group is considered to have been introduced by the Romans, renowned wool users. The Barbarine group, source of the same name breed, is considered as ‘exotic’ by Sagne (1950) because of its Asiatic origin. This breed, the only fat-tailed sheep in Algeria, was introduced at ∼400 BC and was later reintroduced (900 AD) by Arabs from the Near East of Asia (Sarson, 1973). This breed is related to the Tunisian Barbarine and spread throughout the east of the country, from the Oued Souf oasis to the border of Tunisia. Tazegzawt shows a peculiar black-spotted fleece phenotype, with bluish tones observed on muzzle, ears and around the eyes that explains its Kabyle name: the ‘Blue’. This breed, largely ignored until recently and still not mentioned in the FAO DAD-IS (Domestic Animal Diversity Information System of the Food and Agriculture Organization) database (www.fao.org/dad-is), receives increasing attention (FAO, 2014). Indeed, accounting for <0.02% of the Algerian sheep livestock, the situation of this breed, showing high adaptation to mountains of northern Algeria, is critical. Currently, to our knowledge, no document allows making assumptions about the origin of the breed. It may be noted that this breed is reared in the same area as the Berber, the mountains of Kabylie. Contrary to the six other breeds, D’men and Sidaoun do not belong to the ‘wool sheep’ group; D’men falls into the ‘mixed hair-wool sheep’ group and Sidaoun into the ‘hairy sheep’ group.
For each breed, six individuals were sampled, but only five were successfully genotyped for D’men and Barbarine. Individuals were chosen according to their genealogy and collected from multiple flocks (two to four, see Supplementary Table S1 for GPS coordinates) to ensure a representative sampling, except for Hamra that was successfully sampled only in the governmental farm in charge of the breed (Pilot farm of Aïn El Hadjar). Robustness of the sampling design was assessed in Supplementary Text 1 and illustrated by Supplementary Figures S1–S3.
Genotyping and SNP quality control
Blood samples were cryopreserved until DNA extraction and analysis. Genomic DNA was purified from whole blood by protease K digestion and a salting-out procedure (Miller et al., 1988). All animals were genotyped for 54 241 single-nucleotide polymorphism (SNPs) using the Illumina OvineSNP50K Genotyping BeadChip (Illumina, Inc.), following standard operating procedures recommended by the manufacturer. Genotyping was performed by ‘McGill University and Genome Québec innovation center’. SNPs and animals were pruned using the following filtering parameters in order to generate input files: (1) SNP call rate ⩽97%, (2) SNP minor allele frequency ⩽1% and (3) animals displaying ⩾10% of genotype missingness. Quality controls were performed using PLINK v1.07 software (Purcell et al., 2007).
Algerian genotypes were compared with genotypes of European, African, American and Asian sheep populations available from the Sheep HapMap project (Kijas et al., 2012). The genotypes from additional 15 Italian breeds (Ciani et al., 2014) were also included in order to account for possible gene flow between Mediterranean countries. The data set consisted of ∼20 animals per breed, chosen according to their geographic origin, history, census, morphology and production purpose.
Data analysis
Genetic diversity
The level of genetic diversity of Algerian sheep was estimated from the observed percentage of heterozygote genotypes per individual (Ho) and expected heterozygosity (He) using PLINK. The inbreeding coefficients (FIS) were estimated by individual in each breed using PLINK.
Genetic structure
In order to understand the relationship between Algerian breeds, indexes of pair-wise FST of Weir and Cockerham (1984) were determined using ARLEQUIN 3.5 (Excoffier and Lischer, 2010) (permutation procedures were used to derive P-values). A multidimensional scaling analysis was performed using PLINK. To ensure uncorrected linkage disequilibrium did not distort the analysis, SNP pruning was used to identify a SNP subset using the —indep option of PLINK with the following parameters: 50 SNPs per window, a shift of 5 SNPs between windows and a variation inflation factor’s threshold of two (corresponding to r2 >0.5). Pair-wise identical-by-state distance was calculated using all autosomal SNPs that remained after pruning. The graphical representation was depicted using R version 3.0.1 (R Development Core Team, 2013) with RColorBrewer package.
A Bayesian model-based clustering approach was used to search for the occurrence of genetic groups (that is, clusters, K) in our data set (as implemented in STRUCTURE 2.3.4 (Pritchard et al., 2000; Falush et al., 2003, 2007; Hubisz et al., 2009)). Model-based clustering was performed using the admixture model and correlated allele frequencies. The burn-in length of the Markov chain Monte Carlo was set to 50 000 followed by 1 00 000 iterations. Convergence was checked using five runs for each value of K (with K ranging from two to eight, that is, the number of breeds present in the data set). The most probable value of K was estimated by inspection of ΔK statistics (Evanno et al., 2005). The software CLUMPP ver. 1.1.1 (Jakobsson and Rosenberg, 2007) was used to align the repetitions for each K and visualization of results was made by the program DISTRUCT ver. 1.1 (Rosenberg, 2004). Finally, the approach of fineSTRUCTURE (Lawson et al., 2012) based on haplotypes was used. The data set was filtered and phased using SHAPEIT ver. 2 (Delaneau et al., 2012). We used CHROMOPAINTER (Lawson et al., 2012) to analyze the painted data set in order to identify homogenous clusters. Visualization of the posterior distribution of clusters was then performed using the tree-building algorithm of fineSTRUCTURE.
Phylogenetic tree
SNPhylo (Lee et al., 2014), a pipeline specifically developed to construct phylogenetic trees from large SNP data, was used to analyze Algerian data. The pipeline allows for constructing a maximum-likelihood tree with bootstrap values by filtering the SNP data sets using minor allele frequency of 10%; pruning the data set (cutoff threshold 0.8) using SNPRelate (Zheng et al., 2012); constructing multiple alignments of the SNP data set using MUSCLE (Edgar, 2004); determining the maximum-likelihood tree running DNAML from Phylip (Felsenstein, 1989); and performing 1000 bootstraps using Phangorn (Schliep, 2011). The tree file in Newick format provided by the pipeline was processed with MEGA6 (Tamura et al., 2013).
Algerian breeds in an international context
SNPs data from Algerian breeds were merged with available data from world-wide breeds (Kijas et al., 2012) and Italian breeds (Ciani et al., 2014), producing a data set composed of 1625 individuals (a minimum of 3 and a maximum of 20 individuals per breed were considered) from 92 populations (see Supplementary Table S1). A NeighborNet graph, considering all the breeds (except the American breeds BCS, GCN and STE, as they were shown to not cluster with the other American breeds; Kijas et al., 2012) was constructed from a matrix of Reynolds’ distances using Splitstree (Huson, 1998). ADMIXTURE software (Alexander et al., 2009) was used to further investigate relationship between Algerian, Italian American and African sheep breeds. ADMIXTURE was run for K=2 through K=23 (that is, the number of breeds considered), and five independent runs were performed for each value of K. The pophelper R package (Francis, 2016) was used to align, merge and visualize the obtained results. Further analyses were performed with CHROMOPAINTER and fineSTRUCTURE.
Results
Genetic diversity
Considering the 48 Algerian samples analyzed, 1 Barbarine individual failed to produce genotypes and 1 D’men individual showed a call rate of <50%, and the 46 remaining samples showed a call rate of ⩾97%. Out of a total of 54 241 SNPs genotyped, 379 were unmapped and 1450 were located on sex chromosomes, and thus 52 412 SNPs from the 26 sheep autosomes were available and, after filtering (see Materials and methods), 45 755 SNPs passed quality control on 46 individuals from 8 sheep breeds. Allele frequency-dependent diversity estimates are sensitive to ascertainment bias, but the removal of SNP in high linkage disequilibrium acts to counter the effect of the bias, allowing meaningful comparisons among populations (López Herráez et al., 2009). Following linkage disequilibrium-based correction, observed heterozygosity (Ho), calculated by individual, ranged from 0.22 to 0.37, with an average of 0.35 (s.d.=0.030). Average expected heterozygosity (He) was 0.37 (s.d.=0.005). The average FIS value was rather low (mean=−0.07, s.d.=0.09), with 39 individuals showing negative values of FIS. Only three individuals showed pronounced deficit in heterozygosity (two Barbarine and one Tazegzawth) with values of FIS superior to 0.1.
Genetic structure
Calculation of pair-wise FST was used to explore the genetic relationships among breeds. Two groups were detected: (1) Ouled-Djellal, Rembi and Berber showed very low values of pair-wise FST (<0.004), and not significantly different from zero (P<0.001) for Ouled-Djellal/Rembi and Rembi/Berber; and (2) the other breeds showed values of FST ranging from 0.028 (Rembi/Barbarine) to 0.109 (Hamra/Tazegzawt) (Table 1). A total of 4460 pair-wise FST values were calculated considering all together, in the same data set, Algerian breeds, HapMap breeds (Kijas et al., 2012) and Italian breeds (Ciani et al., 2014); the following five lowest pair-wise FST values were observed: Rembi/Ouled-Djellal (0.000); Rembi/Berber (0.002); Ouled-Djellal/Berber (0.004); Australian Merino/Australian Industry Merino (0.004); and Australian Poll Merino/Australian Industry Merino (0.009). All other pair-wise FST values were >0.01 (data not shown).
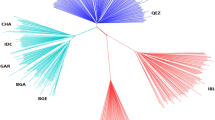
Multidimensional scaling analysis on the matrix of the pair-wise identical-by-state distances among the Algerian sheep samples (Figure 1) showed that Rembi/Ouled-Djellal/Berber were gathered in such a way that breeds cannot be distinguished from one another; Barbarine was close to this ‘group’ but distinct. Tazegzawt and Hamra were definitely distinct from all the other breeds. Sidaoun and D’men were distinguished from the others but close to each other.
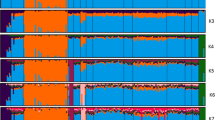
The genetic structure of Algerian sheep breeds was investigated through STRUCTURE analysis, assuming a number of subpopulations (K) in the total sample varying from two to eight (Figure 2). The Tazegzawt was the first breed to be differentiated from all of the others (at K=2). At K=3, a cluster including both Sidaoun and D’men was distinguishable. At K=4 the breed-specific cluster of Hamra appeared. The cluster composed of Ouled-Djellal/Berber/Barbarine/Rembi could not be differentiated before K=5, with individualization of Barbarine starting from this point. At K=8, Berber could be distinguished from Ouled-Djellal and Rembi that were the only breeds remaining still clearly admixed and confounded at this step of the analysis. According to the ΔK criterion (Evanno et al., 2005), the strongest pattern of structuration corresponded to K=2 (ΔK=73.1), highlighting the genetic originality of Tazegzawt. For K=6, a smaller ΔK peak was detected (with ΔK=12.6). This cluster allowed distinguishing between all the breeds except for the group including Berber, Ouled-Djellal and Rembi.
The co-ancestry heatmap (Figure 3) obtained with CHROMOPAINTER/fineSTRUCTURE presents the number of shared genomic ‘chunks’ between the different Algerian individuals. The darker/bluish colors indicate higher co-ancestry estimates, the yellower colors the lower ones. Tazegzawt and Hamra appeared separated from other populations in the number of shared haplotypes. The heatmap showed that Sidaoun and D’men were genetically close to each other. Ouled-Djellal, Rembi and Berber were clearly admixed. Four of the five Barbarine subjects clustered together close to the group formed by Ouled-Djellal/Berber/Rembi, and one Barbarine appeared included into this group. As discussed by Van Dorp et al. (2015), inbreeding can mask signals of ancestry. Hence, the data set was reanalyzed, taking care during the ‘painting’ phase to exclude individuals showing highest values of FIS (seven individuals were excluded from this new analysis). The obtained results (data not shown) were not substantially different, but the three remaining Barbarine individuals appeared included, this time, in the cluster containing Ouled-Djellal and Rembi; moreover, the relatedness between D’men and Sidaoun was more clearly observable.
FineSTRUCTURE clustering for eight Algerian breeds. The color of each bin in the matrix indicates the number of ‘genomic chunks’ copied from a donor (column) to a recipient individual (row). barb, Barbarine; berb, Berber; dmen, D’men; ham, Hamra; OD, Ouled-Djellal; rem, Rembi; sid, Sidaoun; taz, Tazegzawt.
The maximum likelihood tree obtained with SNPhylo (Figure 4) allowed to further examine the genetic relationship among Algerian sheep breeds. As what concerns Tazegzawt, Hamra, Sidaoun and D’men, individuals from the same breed clustered in the same group and for the last two breeds, close genetic relationship was suggested by branch organization. Berber and Rembi individuals were largely admixed together. Barbarine appeared divided into two groups; one group clustering with Rembi individuals and the other clustering with Ouled-Djellal individuals.
Algerian breeds in an international context
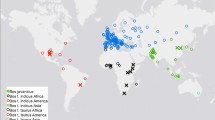
Algerian breeds were also investigated in an international context. A NeighborNet graph, using the Reynolds’ distance and considering Algerian breeds, a subset of breeds from the Sheep HapMap project (see Materials and methods) (Kijas et al., 2012) and Italian breeds (Ciani et al., 2014), was built (Figure 5). Algerian breeds appeared as being at the crossroad of Africa and Europe (Italian breeds, particularly Bagnolese and Laticauda, and, to a lesser extent, with breeds from the Sicily island in Southern Italy: Comisana, Leccese, Valle del Belice and Pinzirita), and also very close to samples from Americas. In order to further investigate these relationships, we adopted the model-based clustering algorithm implemented in the software ADMIXTURE considering a subset from the data set previously mentioned and keeping Algerian breeds, the six breeds from Southern Italy (referred above), the three breeds from Americas and the six African breeds. As our emphasis was on the relationship between Algerian and the other breed groups, we focused on the more explicit clustering solutions for values of K from two to four (Figure 6). At all the tested K values, Algerian breeds highlighted a major (red) component significantly present in Italian breeds. At K=2, they also had a minor (blue) component clearly of African origin. At K=3, the minor (light blue) component was instead mainly shared with breeds from Americas. At K=4, the newly arisen ‘African’ (orange) component appeared in Algerian breeds. Moreover, for K=3 and K=4, clear genetic proximity appeared between Algerian breeds and Barbados Black Belly sheep (BBB, from Caribbean). To refine our perspective on the relationships suggested by the ADMIXTURE analysis, we performed CHROMOPATINER/FineSTRUCTURE analysis (Figure 7). The tree obtained with FineSTRUCTURE confirmed the clear genetic proximity between Algerian and American breeds and highlighted the very peculiar position of Sidaoun showing (1) strong relationship with BBB (from Carribean) and (2) clear genetic proximity with the two Central/Eastern African breeds (Ethiopian Menz (EMZ) and Red Maasai (RMA)). Moreover, the genetic relationship between remaining Algerian breeds (that is, Algerian breeds except Sidaoun) and Italian breeds appeared clearly even if no special affinity for one Italian breed in particular was identified with the analysis.
NeighborNet graph considering Algerian sheep breeds, breeds of the HapMap project (Kijas et al., 2012) and Italian breeds (Ciani et al., 2014) from a matrix of Reynolds’ distances.
FineSTRUCTURE clustering for Algerian, Italian, African and American breeds. The displayed heatmap depicts the number of shared ‘genomic chunks’ between the individuals. Individuals are ordered along each axis according to the inferred tree from fineSTRUCTURE. Afr, Africa; Alg, Algeria; AlgSid, Sidaoun; Am, Americas; Ita, Italy; ItaLec, Leccese.
Discussion
Our comprehensive analysis by genome-wide SNP markers provided original information regarding genetic diversity and history of Algerian sheep breeds.
Diversity and relationships in Algerian sheep
All of the analysis methods adopted, that is, multidimensional scaling analysis (Figure 1), STRUCTURE analysis (Figure 2), fineSTRUCTURE analysis (Figure 3) and the phylogenetic tree (Figure 4), revealed strongly correlated patterns. The main conclusion was that critical situation exists for the Algerian sheep livestock, with a clear dilution of Rembi and Berber occurring mainly through cross-breeding with Ouled-Djellal. Values of pair-wise FST appeared close to zero among these three breeds; also, pair-wise FST considering the world-wide sample showed that the only group with pair-wise FST values as low as the Algerian group (Berber/Ouled-Djellal/Rembi) was the one including three Merino populations from Australia (Australian Industry Merino/Australian Poll Merino/Australian Merino). The Poll Merino and Industry Merino are subtypes of the Australian Merino breed. Hence, Rembi and Berber appeared no more genetically differentiated from Ouled-Djellal than strains of Merino from the Merino breed.
The genetic dilution of the Rembi breed was expected; indeed, cross-breeding practices between Ouled-Djellal and Rembi are denounced for decades (Madani et al., 2003) and, moreover, a previous study from Gaouar et al. (2015) demonstrated using microsatellites markers that Ouled-Djellal and Rembi were highly admixed. From a phenotypic point of view, it was observed that cross-breeding with Ouled-Djellal led to the disappearance of the original Rembi’s phenotype characterized by unicolored bay-fawn fleece (Chellig, 1992) that was replaced by individuals with uniform white fleece resembling Ouled-Djellal (Madani et al., 2003). Berber was analyzed for the first time in the present work. This ancestral breed, considered to be the most ancient breed of the Maghreb, has evolved isolated in Kabylie Mountains during centuries. Berber was accounting for >20% of the Algerian sheep livestock up to 30 years ago, and today it is considered as a ‘minor’ breed (FAO, 2014), indicating important contraction in the livestock population that cannot be rigorously evaluated as no recent census data are available. Sagne (1950) already in 1950 indicated that Berber sheep were more and more replaced by the more productive Ouled-Djellal, and hence admixture could date back to many generations. Barbarine, another very ancient breed, coming from the Near East of Asia, appeared clearly threatened by dilution with the Ouled-Djellal, even if the situation was far from being as critical as that of Rembi and Berber. This breed is the only fat-tail breed of Algeria. All breeds native to the West Asia region have the remarkable ability to store fat in the tail that allows animals to thrive despite fluctuations in feed availability (Iniguez, 2005). The breed is much more represented in Tunisia, where the situation is considered worrying, as intense crosses with the Ouled-Djellal (known there as ‘Western Thin-tailed’) were reported as well as a population decline from 20% between 1964 and 2006 (Aden Aw-Hassan et al., 2008); in Algeria, the Barbarine population has decreased by 60% between 1990 and 2000 (Laaziz, 2005).
The four remaining breeds (Tazegzawt, Hamra, D’men and Sidaoun) appeared not threatened by dilution because of cross-breeding with the Ouled-Djelal. In particular, Tazegzawt and, to a lesser extent, Hamra showed pronounced genetic originality. For these four breeds, the main threat is genetic erosion caused by the high census contraction experienced by the populations that might ultimately lead to their disappearance (Iniguez, 2005). For D’men, in particular, a decrease of 50% was recorded between 1990 and 2000 (Laaziz, 2005). D’men and Sidaoun are Saharan breeds adapted to extreme conditions of drought. D’men falls into the ‘mixed hair-wool sheep’ (Devendra and McLeroy, 1982) and Sidaoun falls into the ‘hairy sheep’ group, and both are thought to be native from West Africa (Mason, 1980). The results of our analysis are in line with this information, pointing to a common origin for these breeds.
Algerian breeds in an international context
Average value of He observed in our study for Algerian sheep was in agreement with those reported by Kijas et al. (2012) and Ciani et al. (2014) in southern and western European breeds using the Illumina OvineSNP50K BeadChip.
The NeighborNet graph (Figure 5) revealed that Algerian breeds were at the crossroads of Africa, Americas and Europe, holding an original position in the world from a genetic point of view. Proximity between Algerian and Italian breeds was observed, particularly with Bagnolese and Laticauda, and, to a lesser extent, with breeds from the Sicily Island (Comisana, Valle del Belice and Pinzirita). Laticauda is the only fat-tailed breed in Italy (Ciani et al., 2014). Together with Bagnolese, the breed originated in Campania (Southern Italy), likely through cross-breeding local sheep from the Apennines with fat-tailed North African sheep, presumably imported under the Bourbons dynasty in the eighteenth century (Ciani et al., 2014). Similarly, Comisana is thought to have been developed at the beginning of the twentieth century in southeast Sicily by cross-breeding local ewes with rams from Malta and North Africa (Ciani et al., 2014). Thus, the observed proximity between Algerian breeds and breeds from Southern Italy would reflect geographical proximity and known exchanges of genetic material.
The particular closeness between Algerian and American sheep breeds (BBB from Caribbean, Morada Nova and Santa Ines from Brazil) (Figures 6 and 7) reflects the transportation of animals during the migration of enslaved West Africans bought to Caribbean and Brazil, starting in the sixteenth century (Ligon, 1657). Masson (1980) suggested an important contribution to D’men (originated from the forest hair sheep of West Africa, and in particular the Fouta Djallon breed of the Cameroon) to the Caribbean BBB sheep. In 1979, Fitzhugh and Bradford (1979) considered, besides the West African origin of the BBB, a possible inclusion of Sahelian sheep in its ancestry. Our results designated the Sidaoun, a Sahel-type sheep, as a major crossover point between African and Caribbean sheep. Further analyses including, among others, the various types of West African sheep breeds are needed to get an even deeper understanding of the Caribbean sheep history.
To conclude, Algerian sheep breeds are in a critical situation, either threatened by genetic dilution or neglected and abandoned. As emphasized by Taberlet et al. (2008), particular attention should be given to the uniqueness of breed features as it is difficult to predict which traits and alleles will be of particular importance in the future.
Data accessibility
Genotypes obtained with the Illumina OvineSNP50K Genotyping BeadChip (genotype files and markers files by chromosomes) for eight Algerian sheep breeds in a PLINK format are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.h0h8d (doi:10.5061/dryad.h0h8d).
References
Aden Aw-Hassan A, Shomo F, Iniguez L . (2008) Small Ruminant Production: Challenges and Opportunities for Poverty Alleviation in West Asia and North Africa. ICARDA: Aleppo, Syria, IV. p 23.
Alexander DH, Novembre J, Lange K . (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664.
Chellig R . (1992) Les Races Ovines Algériennes. Office des Publications Universitaires: Alger. p 80.
Ciani E, Crepaldi P, Nicoloso L, Lasagna E, Sarti FM, Moioli B et al. (2014). Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim Genet 45: 256–266.
Delaneau O, Marchini J, Zagury J-F . (2012). A linear complexity phasing method for thousands of genomes. Nat Methods 9: 179–181.
Devendra C, McLeroy GB . (1982) Goat and Sheep Production in the Tropics. Intermediate Tropical Agriculture Series. Longman Group: UK.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Evanno G, Regnaut S, Goudet J . (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Lischer HEL . (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567.
Falush D, Stephens M, Pritchard JK . (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Falush D, Stephens M, Pritchard JK . (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578.
FAO Mountain Partnership Secretariat, UNCCD, SDC, CDE. (2011) Highlands and Drylands – Mountains, a Source of Resilience in Arid Regions. FAO: Rome.
FAO. (2013) In Vivo Conservation of Animal Genetic Resources. FAO Animal Production and Health Guidelines. No. 14 FAO: Rome.
FAO . (2014). Characterization and value addition to local breeds and their products in the Near East and North Africa – Regional Workshop, Rabat, Morocco, 19–21 November 2012. Animal Production and Health Report No. 3, Rome.
Felsenstein J . (1989). PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166.
Fitzhugh HA, Bradford GE . (1979) Hair sheep production systems a survey of genetic resources. Winrock International Livestock Research and Training Center: Arkansas.
Francis RM . (2016). pophelper: an r package and web app to analyse and visualize population structure. Mol Ecol Resour e-pub ahead of print 6 February 2016doi: 10.1111/1755-0998.12509.
Gaouar SBS, Da Silva A, Ciani E, Kdidi S, Aouissat M, Dhimi L et al. (2015). Admixture and local breed marginalization threaten Algerian sheep diversity. PLoS One 10: e0122667.
Hubisz MJ, Falush D, Stephens M, Pritchard JK . (2009). Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332.
Huson DH . (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics (Oxford, England) 14: 68–73.
Iniguez L . (2005) Characterization of Small Ruminant Breeds in West Asia and North Africa. Vol. 2: North Africa. International Center for Agricultural Research in Dry Areas (ICARDA): Aleppo, Syria.
Jakobsson M, Rosenberg NA . (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806.
Kijas JW, Lenstra JA, Hayes B, Boitard S, Porto Neto LR, San Cristobal M et al. (2012). Genome-wide analysis of the World’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol 10: e1001258.
Laaziz DM . (2005) Small ruminant breeds of Algeria In: Iniguez L (ed.) Characterization of Small Ruminant Breeds in West Asia and North Africa. Vol. 2: North Africa. International Center for Agricultural Research in Dry Areas (ICARDA): Aleppo, Syria.
Lawson DJ, Hellenthal G, Myers S, Falush D . (2012). Inference of population structure using dense haplotype data. PLoS Genet 8: e1002453.
Lee T-H, Guo H, Wang X, Kim C, Paterson AH . (2014). SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15: 162.
Ligon R . (1657) A True and Exact History of the Island of Barbados. Printed for Humphrey Mosely at the Prince's Armes: London.
López Herráez D, Bauchet M, Tang K, Theunert C, Pugach I, Li J et al. (2009). Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs. PLoS One 4: e7888.
Madani T, Yakhlef H, Abbache N . (2003). Les races bovines, ovines, caprines et camelines. In: Abdelguerfi A, Ramdane S (eds) Evaluation des besoins en matière de renforcement des capacités nécessaires à la conservation et l’utilisation durable de la biodiversité importante pour l’agriculture en Algérie, Alger 22-23/01/20013.
MADR DSASI (Ministère de l'Agriculture et du Développement Rural /Direction des Statistiques Agricoles et des Systèmes d’Information) . (2010) Statistiques Élevages, cheptels E2009. (Alger, Algérie.
Mason IL . (1980) Prolific Tropical Sheep FAO Animal Production and Health Paper No. 17. FAO: Rome.
Miller SA, Dykes DD, Polesky HF . (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 12–15.
Pritchard JK, Stephens M, Donnelly P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575.
R Development Core Team . (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna. URL http://www.r-project.org/.
Rosenberg NA . (2004). Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138.
Sagne J . (1950). L'Algérie pastorale. Ses origines, sa formation, son passé, son présent, son avenir. Imprimerie Fontana 27 pp.
Sarson M . (1973). Les ovins dans l’antiquité d’après les vestiges phéniciens et romains en Tunisie et en Algérie. Doc. Tech. INRAT 65.
Schliep KP . (2011). phangorn: phylogenetic analysis in R. Bioinformatics (Oxford, England) 27: 592–593.
Taberlet P, Valentini A, Rezaei HR, Naderi S, Pompanon F, Negrini R et al. (2008). Are cattle, sheep, and goats endangered species? Mol Ecol 17: 275–284.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30: 2725–2729.
Trouette G . (1933) La sélection ovine dans le troupeau indigène. Direction des Services de l'Élevage. Imprimerie P. Guiauchin: Alger.
Turries V . (1976). Les populations ovines algériennes. In: Chaire de zootechnie et de pastoralisme. INA: Alger.
Van Dorp L, Balding D, Myers S, Pagani L, Tyler-Smith C, Bekele E et al. (2015). Evidence for a common origin of blacksmiths and cultivators in the Ethiopian Ari within the last 4500 years: lessons for clustering-based inference. PLoS Genet 11: e1005397.
Weir BS, Cockerham CC . (1984). Estimating F-statistics for the analysis of population-structure. Evolution (NY) 38: 1358–1370.
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS . (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics (Oxford, England) 28: 3326–3328.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
SBS Gaouar: supervised data sampling/wrote the paper; M Lafri: data sampling/wrote the paper; A Djaout, R El-Bouyahiaoui, A Bouri and A Bouchatal: acquisition of data; A Maftah: designed the research; E Ciani: wrote the paper; A Da Silva: designed the research/analyzed data/wrote the paper.
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Gaouar, S., Lafri, M., Djaout, A. et al. Genome-wide analysis highlights genetic dilution in Algerian sheep. Heredity 118, 293–301 (2017). https://doi.org/10.1038/hdy.2016.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.86
This article is cited by
-
Genome variation in tick infestation and cryptic divergence in Tunisian indigenous sheep
BMC Genomics (2022)
-
Genome-wide insights on gastrointestinal nematode resistance in autochthonous Tunisian sheep
Scientific Reports (2021)
-
Genetic homogenization of indigenous sheep breeds in Northwest Africa
Scientific Reports (2019)
-
Genomic diversity and population structure of three autochthonous Greek sheep breeds assessed with genome-wide DNA arrays
Molecular Genetics and Genomics (2018)