Abstract
The endophilic tick Ixodes arboricola infests cavity-nesting birds, and its dispersal strongly depends on the movements of its host. Population genetic structure of I. arboricola was studied with seven polymorphic microsatellite markers. We collected 268 ticks from 76 nest boxes in four woodlots near Antwerp, Belgium. These nest boxes are mainly used by the principal hosts of I. arboricola, the great tit Parus major and the blue tit Cyanistes caeruleus. As these birds typically return to the same cavity for roosting or breeding, ticks within nest boxes were expected to be highly related, and tick populations were expected to be spatially structured among woodlots and among nest boxes within woodlots. In line with the expectations, genetic population structure was found among woodlots and among nest boxes within woodlots. Surprisingly, there was considerable genetic variation among ticks within nest boxes. This could be explained by continuous gene flow from ticks from nearby tree holes, yet this remains to be tested. A pairwise relatedness analysis conducted for all pairs of ticks within nest boxes showed that relatedness among larvae was much higher than among later instars, which suggests that larvae are the most important instar for tick dispersal. Overall, tick populations at the studied spatial scale are not as differentiated as predicted, which may influence the scale at which host–parasite evolution occurs.
Similar content being viewed by others
Introduction
Dispersal of parasites and their hosts is one of the most important factors affecting the dynamics and coevolution of host–parasite interactions (Gandon et al., 1996; Clayton and Moore, 1997). Parasite population structure is often tightly linked to host dispersal, because parasites are typically less mobile than their hosts (Boulinier et al., 2001). If parasites are unable to make use of their hosts’ dispersal capabilities to a full extent, parasite populations may be vulnerable to inbreeding and have reduced coevolutionary potential (Clayton and Moore, 1997).
Most ixodid ticks have low intrinsic dispersal capacities and therefore depend on host movement for transportation over large distances (Falco and Fish, 1991). For instance, the seabird tick Ixodes uriae can disperse autonomously to nearby bird nests but needs the host to colonise new breeding cliffs. As a consequence, the spatial arrangement of the host determines the population genetic structure of the ticks (McCoy et al., 2003). Ixodid ticks spend most of their time off-host because all instars (that is, larvae, nymphs, adult females but not adult males) typically take a single blood meal lasting several days before detaching from the host and moulting to the next instar. To survive and find a new host, ticks must detach in suitable habitat. Hence, tick species with different ecological requirements employ different detachment strategies, even if they infest the same host species (Heylen and Matthysen, 2010).
Endophilic ticks remain hidden close to their hosts’ nest or burrow and attach when the host arrives (Salman and Tarrés-Call, 2012). Whereas this maximises the possibility of finding a host, it limits dispersal among nests because these are discrete habitats in a matrix of unsuitable habitat that is difficult to cross autonomously. Furthermore, if hosts show high nest fidelity ticks will tend to detach in the same nest (McCoy et al., 2003). Thus the population genetic structure of endophilic ticks is expected to depend on the host’s ecology, though parasite gene flow can be much lower than expected based on host mobility.
In this study, the population genetic structure of the endophilic tick Ixodes arboricola Schulze and Schottke was investigated near Antwerp (Belgium) to gain insight in the transmission ecology of a host–parasite system of which little is known (Hillyard, 1996). I. arboricola infests mainly cavity-nesting birds (mostly great tits Parus major and blue tits Cyanistes caeruleus) (Hudde and Walter, 1988), shows a strong tendency to detach within cavities (Heylen and Matthysen, 2010; White et al., 2012) and infests birds typically from autumn to late spring (Heylen et al., 2014). The feeding activity of larvae increases in late autumn, and the majority of feeding nymphs and adult females can be found during the birds’ breeding season, when adult female ticks primarily infest nestlings (Heylen et al., 2014). Whereas female ticks feed in the adult instar, male ticks feed only in the larval and nymphal stages and remain in the host’s cavity in the adult stage, where they copulate with both unfed and engorged female ticks (Heylen et al., 2012).
Although great and blue tits are intrinsically very mobile, can disperse over large distances and even may show seasonal migration (Gosler, 1993), ticks can only disperse by feeding on birds that move from one cavity to another before the ticks detach. Great and blue tits mainly disperse during the first summer and autumn after leaving their natal nest box (Dhondt, 1979), but they rarely sleep in cavities in summer and early autumn (Gosler, 1993). In the breeding season, birds occupy only a single cavity to raise their young (Gosler, 1993) so that dispersal opportunities are restricted to birds that produce a second clutch in a different cavity, but the frequency of second clutches is very low in woodlots near Antwerp (Matthysen et al., 2011). Finally, once birds have established a territory they tend to return to this area for breeding and roosting throughout their life (Gosler, 1993). Ticks may therefore disperse mainly when birds prospect cavities to find a suitable breeding location. In winter, birds may also switch among different roosting sites, although they tend to return to the same box (Tyller et al., 2012). Hence, opportunities for I. arboricola to disperse among cavities are limited, which should affect population genetic structure.
Because of the spatial ecology of their hosts, it was expected that I. arboricola has limited dispersal opportunities and therefore displays spatial genetic structure, that is, genetic differentiation and patterns of isolation by distance among woodlots and among nest boxes within woodlots. In addition, levels of relatedness among ticks within nest boxes should be high. Nevertheless, even limited dispersal opportunities were expected to lead to genetic differentiation among different tick instars within nest boxes, because they are different generations (cohorts) that may disperse among nest boxes. Finally, it was expected that pairwise relatedness among ticks within nest boxes depends on their developmental stage, because ticks have a single opportunity to disperse with every feeding bout. More specifically, it was expected that, on average, the relatedness of pairs of individuals that have a higher sum of past feeding bouts is lower than that of pairs of individuals with a lower sum of past feeding bouts. The present study aims at testing these expectations by means of a survey of allelic variation at seven polymorphic microsatellite loci in I. arboricola from woodlots in the vicinity of Antwerp, Belgium.
Methods
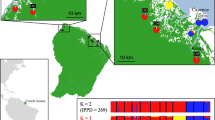
I. arboricola ticks used in the current study were sampled from four woodlots near Antwerp, Belgium between December 2010 and March 2013: Peerdsbos, Brasschaat (PB), Middelheim, Antwerp (MI), De Warande, Oostmalle (WA), and De Kolonie, Wortel (WK) (Figures 1 and 2, Table 1). Ticks were discovered in PB in 2006, a woodlot subject to long-term population studies on great and blue tits for many years (Matthysen et al., 2011). Ticks were subsequently found in MI, WA and WK during a survey of >1000 nest boxes (used mainly by great and blue tits) in nine woodlots throughout Belgium in 2010 and 2011. No, or very few, I. arboricola ticks have been found in the remaining woodlots.
Estimated population structure. The most likely number of populations (K) was estimated with a Bayesian approach for each woodlot separately. Each bar represents an individual tick, and the number below each bar indicates its developmental stage (1=unfed larvae; 2=engorged larvae and unfed nymphs; 3=engorged nymphs, adult males and unfed adult females; and 4=engorged adult females). Ticks are clustered per nest box, and labels above each graph represent sampling dates (month+year). (a) MI: 15 nest boxes (of which four were sampled twice); (b) PB: 25 nest boxes (of which one was sampled twice); (c) WA: 20 nest boxes (of which one was sampled twice); (d) WK: 16 nest boxes (none sampled twice).
Ticks of all instars (eggs, larvae, nymphs and imagos; both unfed and engorged) were collected and stored in 70–100% ethanol. Species, developmental stage, sex (only for adults) and engorgement status were identified using a Leica MZ125 stereomicroscope (Leica Microsystems, Wetzlar, Germany) and identification keys (Arthur, 1963; Hillyard, 1996). In total, 268 I. arboricola ticks (excluding eggs) were collected from 76 nest boxes, and the number of ticks analysed per nest box ranged from 1 to 11 (Table 1). In six nest boxes, ticks were collected on two occasions: in MI four nest boxes were sampled in June 2011 (yielding 8, 8, 2 and 4 ticks, respectively) and June 2012 (yielding 1, 10, 2 and 6 ticks, respectively). In PB, one tick was collected from a nest box in December 2010 and one from the same nest box in May 2012. In WA, five ticks were acquired from a nest box in June 2011 and six from the same nest box in March 2013. These numbers are too low to test for genetic differentiation over time. To prevent pseudoreplication, the second sampling events were not used in any analysis except the Bayesian clustering analysis (see below).
Using NucleoSpin Tissue kits (Macherey-Nagel, Düren, Germany), DNA was extracted from whole individual larvae and nymphs and from three legs in case of adults. The first pair of legs was never used, as these are important for species identification. Ten polymorphic microsatellite loci were amplified following Van Houtte et al. (2013) and analysed with a 3130XL GENETIC ANALYSER (Life Technologies, Carlsbad, CA, USA). Alleles were scored with GENEMAPPER VERSION 3.7 (Life Technologies). In this way, it was possible to genotype 253 I. arboricola ticks, with 192 ticks yielding successful amplification at all loci and 61 ticks yielding only partial success. In 15 ticks, none of the microsatellites amplified successfully.
Genetic variability in the four woodlots was assessed with GENEPOP VERSION 4.0 at the woodlot-level (Rousset 2008, Mol Ecol Resour). This software was used to estimate observed (HO) and expected (HE) heterozygosity, the inbreeding coefficient (FIS) and the frequency of null alleles, test for linkage disequilibrium between pairs of loci in each woodlot and test for deviations of genotype frequencies from Hardy-Weinberg equilibrium (HWE) expectations using an exact probability test. Following Chapuis and Estoup (2007), microsatellite loci that showed significant heterozygote deficiencies (HDs, that is, positive FIS) at all woodlots were discarded from all analyses, because this is indicative of null alleles, and the presence of null alleles may lead to overestimation of genetic differentiation (Chapuis and Estoup, 2007). At the nest box-level, tests for deviations from HWE expectations were conducted for nest boxes with >5 individuals using exact probability tests across all loci. The sequential Bonferroni’s procedure was applied to the tests of linkage disequilibrium and FIS to correct for multiple testing (Rice, 1989).
The population genetic structure of I. arboricola was investigated in several ways. First, isolation by distance was tested among all nest boxes combined and among nest boxes within woodlots for every woodlot separately by comparing a pairwise log-transformed geographic distance matrix to a pairwise genetic distance matrix with a Mantel test (1000 permutations) in GENEPOP VERSION 4.0 (Rousset, 2008). Pairwise geographic distances were calculated with the DIST-function in R VERSION 3.0.1 using Decimal Degree coordinates (R Core Team, 2013), and pairwise genetic distances were calculated as FST/(1−FST) in GENEPOP VERSION 4.0 (Rousset, 2008).
Second, an analysis of molecular variance (AMOVA), executed in ARLEQUIN VERSION 3.11 (Excoffier et al., 2005), was used to partition genetic variation at three levels, namely, among woodlots, among nest boxes within woodlots and within nest boxes. Because of the software’s inability to use loci with too many missing data, the analysis was restricted to the 192 ticks for which all microsatellites were scored successfully. Because different instars within nest boxes represent different generations (cohorts), AMOVA was also used to investigate genetic differentiation among tick instars (that is, cohorts) within nest boxes by partitioning genetic variation at three levels, namely, among nest boxes, among cohorts within nest boxes and within cohorts. All nest boxes were included in this analysis, but variation among cohorts within nest boxes could only be estimated from 29 nest boxes where several cohorts were collected.
Third, a Bayesian approach for inferring population genetic structure among nest boxes was applied in STRUCTURE VERSION 2.3.4 (Falush et al., 2003). For every woodlot, the number of assumed populations (K) was simulated from one to the number of sampled nest boxes with 20 replications, where doubly sampled nest boxes were included twice (KMAX: MI=19, PB=26, WK=21, WA=16). For all runs (50 000 MCMC repeats with a burn-in of 5 000), the admixture model was implemented, which assumes that individuals have derived ancestry from more than one population, as well as the correlated-allele-frequencies model, which calculates allele frequencies over all populations. The software STRUCTURE HARVESTER VERSION 0.6.93 (Earl et al., 2012) was used to select the most likely number of populations for every woodlot, that is, K with the highest value of Delta K. For this K, the replication from the software STRUCTURE VERSION 2.3.4 with the highest likelihood was selected for graphical presentation. For every tick, the proportion of the genome originating from each inferred population was computed (quantitative clustering method).
Finally, the expectation was tested that relatedness among individual ticks in a nest box differs among instars and decreases with the number of dispersal opportunities separating them since the larval stage. Because ticks feed once per instar and subsequently moult to the next instar, every feeding bout is a dispersal opportunity, namely, (1) unfed larvae have never fed and therefore had zero dispersal opportunities, (2) engorged larvae moult into unfed nymphs and both have had one dispersal opportunity, (3) engorged nymphs, and the adults they moult into, have had two dispersal opportunities, and (4) adult males do not feed but adult female ticks do, so that males and unfed females have had two, and engorged females have had three dispersal opportunities. The maximum number of dispersal events of a pair of individuals is therefore the sum of dispersal opportunities of both individuals.
The sum of dispersal opportunities was calculated for every pair of individuals within nest boxes. Ticks from resampled nest boxes (26 ticks from six nest boxes) were discarded, and nest boxes with only one individual (30 nest boxes but only 28 individuals because two individuals coming from resampled nest boxes were already discarded) could not be used, hence 199 individuals from 46 nest boxes were used, representing 421 comparisons. There were seven categories, comprising 0–6 dispersal opportunities (Table 2). The program SPAGEDI VERSION 1.4a (Hardy and Vekemans 2002, Mol Ecol Notes) was used to calculate relatedness (r) for every pair of individuals and conduct linear and logarithmic regression analyses across pairs of individuals within nest boxes, testing the association between r and the sum of dispersal opportunities. Queller and Goodnight (1989) estimator of relatedness was used, which takes allele frequencies into account, eliminates a downward bias for small sample sizes and allows estimation of relatedness for subsets of population samples. Negative kinship coefficients indicate that two individuals are less related than random individuals (Hardy and Vekemans, 2002). The regression analyses were conducted with 10 000 permutations over individuals and loci.
Results
Three microsatellite loci (Ixaf17, Ixaf18 and Ixaf20) were discarded, because FIS was significantly positive in all populations for these loci. Consequently, all analyses were conducted with the remaining seven loci (Table 1). The discarded loci had a mean frequency of null alleles across all woodlots of >15% (Ixaf17: 18.4%; Ixaf18: 30.1%; Ixaf20: 18.8%), whereas the mean frequency of null alleles across all woodlots for the remaining seven microsatellite loci was lower (Ixaf3: 5.2%; Ixaf6: 7.4%; Ixaf8: 11.1%; Ixaf11: 5.6%; Ixaf15: 4.9%; Ixaf16: 9.4%; Ixaf19: 11.1%). The HO, HE and FIS values and frequencies of null alleles per population–locus combination of the remaining seven microsatellite loci are given in Table 1. After Bonferroni’s correction, 10 out of the 28 population–locus combinations deviated significantly from HWE expectations. These deviations were observed in all populations and at all loci except Ixaf15. There was no significant linkage disequilibrium. At the nest box-level, there were no deviations from HWE expectations (Table 3).
The AMOVA of population genetic structure within and among cohorts showed significant genetic structure among nest boxes (Φnest boxes−total=0.14, df=73, P<0.001, variation explained: 10.9%) and within cohorts (Φcohorts=0.11, df=264, P<0.001, variation explained: 86.3%) but not among cohorts within nest boxes (Φcohorts−nest boxes=0.03, df=22, P=0.150, variation explained: 2.8%).
There was no significant isolation by distance, neither in the overall test (R2=0.003, P=0.15) nor in any of the woodlots (MI: R2=0.004, P=0.31; PB: R2=0.001, P=0.49; WA: R2=0.011, P=0.16; WK: R2=0.028, P=0.07). The AMOVA of population genetic structure within and among woodlots showed significant genetic differentiation at all investigated levels, namely, among woodlots, among nest boxes within woodlots and within nest boxes (Table 4).Within woodlots, the Bayesian clustering analysis resulted in K=3 for MI, K=5 for PB, K=2 for WA and K=4 for WK (Figure 2). In MI, ticks in five nest boxes, (d [1+2], h, l, n, o) were assigned to the first cluster, ticks in six nest boxes (a, b [1+2], c [1+2], e [2], f, g, i, j) to the second and ticks in two nest boxes (k, m) to the third cluster. Ticks in two nest boxes (e [1]) were not clearly assigned to any of the clusters. In PB, ticks in most nest boxes were not assigned to particular clusters. Ticks in five nest boxes (a [1+2], i, k, l, o) were assigned to the first cluster. In WA, all ticks were equally assigned to both clusters. In WK, four clusters were identified, and ticks from the nest boxes sampled in November 2011, except nest box f, were assigned to clusters 1–3, whereas ticks from the single nest box sampled in April 2012 were assigned to the fourth cluster.
Relatedness decreased significantly with an increasing sum of dispersal opportunities when the latter was logarithmically transformed (b=−0.173; R2=0.050; P=0.022; Figure 3) but not when it was linear (b=−0.039; R2=0.029; P=0.099). The mean r for the categories with 0, 1 and 2 dispersal opportunities was higher (mean: 0.210–0.359; range: −0.853 to 1.000) than the mean r for the categories with 3–6 dispersal opportunities (mean: 0.053–0.128; range: −1.895 to 1.000).
Queller and Goodnight (1989) estimator of relatedness (QGE) for pairwise relatedness between ticks within nest boxes. Relatedness decreases significantly with increasing dispersal opportunities (b=−0.173; R2=0.050; P=0.022). Data are categorised by the number of dispersal opportunities separating individual ticks. Presented here is QGE based on individual pairs, whereas the analysis was conducted on global estimates.
Discussion
Knowledge concerning population genetic structure is essential for the understanding of parasite dispersal and the evolution of host specificity and coevolution in host–tick interactions (Boulinier et al., 2001; McCoy et al., 2003). The current study is the first that investigated spatial genetic structure of a bird-specialised tick in a terrestrial ecosystem, following work on other endophilic ticks (McCoy et al., 2003; Guzinski et al., 2009; Dharmarajan et al., 2011). Because I. arboricola has a low intrinsic mobility (Heylen and Matthysen, 2010) while its main hosts (P. major and C. caeruleus) show high fidelity to cavities (Gosler, 1993), tick populations were expected to be spatially structured among woodlots and among nest boxes within woodlots and ticks within nest boxes to be highly related.
In line with the expectations, tick populations were significantly differentiated among the four woodlots and among nest boxes within woodlots. At woodlot-level, several population–locus combinations showed significant HDs. HDs can be caused by methodological (for example, null alleles) and biological (for example, population subdivision, inbreeding) factors. It is generally believed that null alleles lead to locus-specific effects, whereas biological factors lead to a more or less concordant pattern across all loci (Dakin and Avise, 2004). Biological and methodological factors can only be disentangled through experimental testing and, unfortunately, there is no methodology to accommodate for null alleles in population genetic analyses (unless for the estimation of genealogical relationships, see, for example, Kalinowski and Taper (2006) and, for FST-values, see Chapuis and Estoup (2007)). The presence of null alleles may have led to overestimation of FIS and genetic differentiation, which could indicate tick populations are not as differentiated as the current data suggest (Chapuis and Estoup, 2007). On the other hand, no locus-specific effects were found for the seven microsatellite loci used in the current study. Therefore HDs in the current study may have been caused by biological factors, and null allele frequencies may have been overestimated. The fact that significant genetic differentiation was found among nest boxes within woodlots suggests that HDs are caused by subdivision of tick populations among nest boxes within woodlots (Wahlund effect). This is in line with studies of the raccoon tick I. texanus, where highly variable levels of HD among loci and populations were caused by subdivided breeding groups and high variance in individual reproductive success (Dharmarajan et al., 2011). The fact that no HDs have been found within nest boxes supports the claim that tick populations are subdivided within woodlots, although power of this test is low and hence further work is needed to draw more decisive conclusions.
Against expectations, there was considerable genetic variation among ticks within nest boxes. This may explain the lack of genetic differentiation among cohorts within nest boxes. High levels of genetic variation among ticks within nest boxes suggest that not all genotypes have been sampled, especially as the number of ticks collected from individual nest boxes is limited. It is known that I. arboricola ticks can be easily overlooked during nest box surveys, especially the small unfed immature instars (Heylen et al., 2014). Nevertheless, because all nest boxes in a woodlot were inspected during nest box surveys (except during III 2013 in WA and IV 2012 in WK), the majority of ticks present in a woodlot will have been collected, and ticks may come from outside the nest boxes in the woodlots. No sampling was conducted in between the four woodlots, because there are no major nest box sites, but ticks may be present in nest boxes and other cavities used by birds in gardens and parks. These cavities are probably very important for tick dispersal, because great and blue tits are unlikely to transfer ticks directly among woodlots that are separated by several kilometres (Gosler, 1993). This corroborates with the fact that significant genetic differentiation was found among woodlots but would also result in isolation by distance among woodlots. Further investigations are required to assess whether ticks are able to disperse among woodlots that are separated by several kilometres. We expect the large genetic differentiation within nest boxes, low sample size and incomplete gene pool are responsible for the lack of isolation by distance among woodlots.
One hypothesis for the high within-nest differentiation is that there is inflow of ticks from natural cavities (that is, tree holes), which are abundant in woodlots such as the studied ones (Bai, 2005). Although great and blue tits have a preference for nest boxes, tree holes may be visited when birds change winter roosting sites, inspect cavities in spring and for breeding (Lohmus and Remm, 2005). Tree holes may provide not only additional but also higher-quality habitat to I. arboricola, because they have more cracks and crevices that provide hiding places for ticks and probably have higher humidity, to which ticks are sensitive (Sonenshine, 1991). It is difficult to collect ticks from tree holes, and the prevalence of ticks within tree holes remains to be investigated. If tree holes are indeed more suitable habitat for I. arboricola, they may have higher population densities than nest boxes and result in a sink-source system with asymmetrical gene flow (Dias, 1996).
In line with the idea that nest boxes are unsuitable habitat for I. arboricola, woodlots that were surveyed once yielded very few ticks and I. arboricola seems to be rare in nest boxes throughout Belgium. However, the abundance of ticks was high in WA, where nest boxes are much older, more moist and contain many more cracks and crevices than in the other woodlots (unpublished data). Such nest boxes might, like tree holes, be more suitable for I. arboricola and be the reason tick abundance is considerably higher in WA than in other woodlots. Interestingly, WA is the only woodlot where nest boxes do not explain the genetic structure that was found, but the current data do not allow to investigate whether there is a relationship between tick abundance and genetic structure within woodlots. It might be worthwhile to study WA in more detail, because this woodlot might be a close approximation to a natural population of I. arboricola. This might also provide more insight in the temporal characteristics of I. arboricola populations. Although some nest boxes were sampled successively in the current study, the sample is too small to make reliable predictions about temporal variation in genetic structure of I. arboricola populations. In addition, in many cases all ticks were collected, because tick abundance was low, and therefore successive samples may be completely unrelated.
Despite the considerable genetic differentiation within nest boxes, relatedness decreased with increasing dispersal opportunities, supporting the hypothesis that feeding bouts are indeed dispersal events. However, instead of decreasing gradually, there was a sudden drop in relatedness after two dispersal opportunities, suggesting that different tick instars may have different dispersal opportunities. Most engorged larvae are found in autumn, when birds may switch between roosting sites, to early spring, when birds inspect possible nesting sites (Gosler, 1993), whereas engorged nymphs and adult females are mainly found during the birds’ breeding season, when birds show high nest fidelity (Heylen et al., 2014). Together with the current data, this suggests that the majority of dispersal occurs in autumn, in the early life of ticks. Interestingly, there was a lot of variation in relatedness among pairs of ticks, and some pairs of unfed larvae displayed very low relatedness. It is possible that unfed larvae were sampled that came from clutches laid by different females. Furthermore, larvae from the same clutch might be fathered by multiple males, as is the case in other ixodid ticks (McCoy and Tirard, 2002; Ruiz-Lopez et al., 2012). The mating strategies of I. arboricola are largely unknown as of yet, and whether dispersal is indeed distributed unequally among instars needs further investigation.
An unknown factor in the current study is the degree to which ticks have all originated from the same host species. Although there is no exact data on birds using the nest boxes in the period of sampling, most ticks were assumed to have fed on great and blue tits. Great and blue tits are known from regular inspections in PB and observations from bird ringers from the other woodlots to be the main occupants of nest boxes. Occasionally, nest boxes are occupied by other songbirds, notably nuthatches (Sitta europaea) and pied flycatchers (Ficedula hypoleuca). In both of these species, parasitism by I. arboricola has been described earlier (Hudde and Walter, 1988). Nuthatches and pied flycatchers greatly overlap with great and blue tits in nest-site selection and can therefore be expected to contribute as hosts to the same I. arboricola populations feeding on great and blue tits. Interestingly, there are occasional reports that I. arboricola also parasitises hole-nesting birds that differ more strongly in nest use from great and blue tits, such as owls and jackdaws (Hudde and Walter, 1988; Hillyard, 1996). Tick dispersal among different nest types may be limited, and consequently, tick populations among hosts with different nesting requirements may be genetically differentiated. At this point, however, there can be only speculation about this as I. arboricola has not been found in such nests in Belgium as of yet (unpublished data).
In conclusion, this is the first population genetic study of an ornithophilic tick in a terrestrial system. Genetic differentiation was found among the studied woodlots and among nest boxes within woodlots. Contrary to expectations, genetic differentiation was found among individuals within nest boxes. Relatedness analyses showed that dispersal is unequal among instars, and larvae seem the most important instar for dispersal of I. arboricola.
Data archiving
Genotype data have been submitted to Dryad: doi:10.5061/dryad.hb169.
References
Arthur DR . (1963) British Ticks. Butterworths: London, UK.
Bai ML . (2005) Tree cavity abundance and nest site selection of cavity nesting birds in a natural boreal forest of West Khentey, Mongolia. PhD thesis. Georg-August-Universität: Göttingen, Germany.
Boulinier T, McCoy KD, Sorci G . (2001). Dispersal and parasitism. In: Clobert J, Danchin E, Dhondt A, Nichols JD, (eds). Dispersal. Oxford University Press: Oxford, UK. pp 169–179.
Chapuis MP, Estoup A . (2007). Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24: 621–631.
Clayton DH, Moore J . (1997) Host-Parasite Evolution: General Principles and Avian Models. Oxford University Press: Oxford, UK.
Dakin EE, Avise JC . (2004). Microsatellite null alleles in parentage analysis. Heredity 93: 504–509.
Dharmarajan G, Beasley JC, Rhodes OE Jr. . (2011). Heterozygote deficiencies in parasite populations: an evaluation of interrelated hypotheses in the raccoon tick, Ixodes texanus. Heredity 106: 253–260.
Dhondt AA . (1979). Summer dispersal and survival of juvenile great tits in southern Sweden. Oecologia 42: 139–157.
Dias PC . (1996). Sources and sinks in population biology. Trends Ecol Evol 11: 326–330.
Earl DA, vonHoldt BM, Bridget M . (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361.
Excoffier L, Laval G, Schneider S . (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50.
Falco RC, Fish D . (1991). Horizontal movement of adult Ixodes dammini (Acari, Ixodidae) attracted to CO2-baited traps. J Med Entomol 28: 726–729.
Falush D, Stephens M, Pritchard JK . (2003). Inference of population structure: Extensions to linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I . (1996). Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc R Soc Lond Ser B-Biol Sci 263: 1003–1009.
Gosler A . (1993) The Great Tit. Hamlyn: London, UK.
Guzinski J, Bull CM, Donnellan SC, Gardner MG . (2009). Molecular genetic data provide support for a model of transmission dynamics in an Australian reptile tick Bothriocroton hydrosauri. Mol Ecol 18: 227–234.
Hardy OJ, Vekemans X . (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Heylen DJA, Matthysen E . (2010). Contrasting detachment strategies in two congeneric ticks (Ixodidae) parasitizing the same songbird. Parasitology 137: 661–667.
Heylen DJA, Van Oosten AR, Devriendt N, Elst J, Jacobs I, De Bruyn L et al. (2014). Seasonal feeding activity of the tree-hole tick, Ixodes arboricola. Parasitology (e-pub ahead of print 21 March 2014).
Heylen DJA, White J, Elst J, Jacobs I, van de Sande C, Matthysen E . (2012). Nestling development and the timing of tick attachments. Parasitology 139: 766–773.
Hillyard PD . (1996) Ticks of North-West Europe: keys and notes for identification of the species. Published for the Linnean Society of London and the Estuarine and Coastal Sciences Association by Field Studies Council.
Hudde H, Walter G . (1988). Verbreitung und Wirtswahl der Vogelzecke Ixodes arboricola (Ixodoidea, Ixodidae) in der Bundesrepublik Deutschland. Vogelwarte 34: 201–207.
Kalinowski S, Taper M . (2006). Maximum likelihood estimation of the frequency of null alleles at microsatellite loci. Conserv Genet 7: 991–995.
Lohmus A, Remm J . (2005). Nest quality limits the number of hole-nesting passerines in their natural cavity-rich habitat. Acta Oecol 27: 125–128.
Matthysen E, Adriaensen F, Dhondt AA . (2011). Multiple responses to increasing spring temperatures in the breeding cycle of blue and great tits (Cyanistes caeruleus, Parus major). Glob Change Biol 17: 1–16.
McCoy KD, Tirard C . (2002). Reproductive strategies of the seabird tick Ixodes uriae (Acari: Ixodidae). J Parasitol 88: 813–816.
McCoy KD, Tirard C, Michalakis Y . (2003). Spatial genetic structure of the ectoparasite Ixodes uriae within breeding cliffs of its colonial seabird host. Heredity 91: 422–429.
Queller DC, Goodnight KF . (1989). Estimating relatedness using genetic markers. Evolution 43: 258–275.
R Core Team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Rice WR . (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Rousset F . (2008). Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8: 103–106.
Ruiz-Lopez MJ, Chaskelson S, Gompper ME, Eggert LS . (2012). Multiple paternity in the American dog tick Dermacentor variabilis (Acari: Ixodidae). J Parasitol 98: 498–501.
Salman MD, Tarrés-Call J . (2012) Ticks and Tick-Borne Diseases: Geographical Distribution and Control Strategies in the Euro-Asia Region. CAB International: Wallingford, UK.
Sonenshine DE . (1991) Biology of Ticks. Oxford University Press: Oxford, UK.
Tyller Z, Paclík M, Remes V . (2012). Winter night inspections of nest boxes affect their occupancy and reuse for roosting by cavity nesting birds. Acta Ornithol 47: 79–85.
Van Houtte N, Van Oosten AR, Jordaens K, Matthysen E, Backeljau T, Heylen DJ . (2013). Isolation and characterization of ten polymorphic microsatellite loci in Ixodes arboricola, and cross-amplification in three other Ixodes species. Exp Appl Acarol 61: 327–336.
White J, Heylen DJA, Matthysen E . (2012). Adaptive timing of detachment in a tick parasitizing hole-nesting birds. Parasitology 139: 264–270.
Acknowledgements
We thank I Jacobs and J Elst for field assistance and N Van Houtte and K Breugelmans for laboratory assistance. In addition, we thank three anonymous referees for comments on a previous draft of this manuscript. This study was funded by FWO-project G.0049.10 to EM and TB. DJAH is a postdoctoral fellow at FWO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Van Oosten, A., Heylen, D., Jordaens, K. et al. Population genetic structure of the tree-hole tick Ixodes arboricola (Acari: Ixodidae) at different spatial scales. Heredity 113, 408–415 (2014). https://doi.org/10.1038/hdy.2014.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.41
This article is cited by
-
An experimental test to compare potential and realised specificity in ticks with different ecologies
Evolutionary Ecology (2016)