Abstract
Maize Abnormal chromosome 10 (Ab10) contains a classic meiotic drive system that exploits the asymmetry of meiosis to preferentially transmit itself and other chromosomes containing specialized heterochromatic regions called knobs. The structure and diversity of the Ab10 meiotic drive haplotype is poorly understood. We developed a bacterial artificial chromosome (BAC) library from an Ab10 line and used the data to develop sequence-based markers, focusing on the proximal portion of the haplotype that shows partial homology to normal chromosome 10. These molecular and additional cytological data demonstrate that two previously identified Ab10 variants (Ab10-I and Ab10-II) share a common origin. Dominant PCR markers were used with fluorescence in situ hybridization to assay 160 diverse teosinte and maize landrace populations from across the Americas, resulting in the identification of a previously unknown but prevalent form of Ab10 (Ab10-III). We find that Ab10 occurs in at least 75% of teosinte populations at a mean frequency of 15%. Ab10 was also found in 13% of the maize landraces, but does not appear to be fixed in any wild or cultivated population. Quantitative analyses suggest that the abundance and distribution of Ab10 is governed by a complex combination of intrinsic fitness effects as well as extrinsic environmental variability.
Similar content being viewed by others
Introduction
Major deviations from Mendel’s rules have been termed meiotic drive, signifying preferential transmission of chromosomes or chromosome regions to progeny (Sandler and Novitski, 1957). Although deviations in meiosis are implied by the term, the most heavily studied examples of meiotic drive affect post-meiotic events. Three well-characterized examples are the t-haplotype in Mus musculus, segregation distorter in Drosophila melanogaster, and spore killer in Neurospora intermedia (Burt and Trivers, 2006). In these examples meiotic drive is controlled by multiple loci that encode trans-acting factors. The loci involved are tightly linked in a haplotype shielded from recombination by genomic rearrangements and/or proximity to pericentromeric heterochromatin (Lyttle, 1991; Burt and Trivers, 2006). In all three cases the mechanism of meiotic drive involves disruption of sperm or spore sterility (Lyttle, 1991; Burt and Trivers, 2006).
In contrast, the maize abnormal chromosome 10 (Ab10) meiotic drive system changes meiosis in a way that results in the preferential inclusion of heterochromatin-based knobs in reproductive cells (Rhoades, 1952). Ab10 contains an extended chromatin region at the end of the long arm roughly the size of the short arm of a normal chromosome 10 (Hiatt and Dawe 2003a). This region is a complex haplotype consisting of at least three knobs and several genes (Mroczek et al., 2006) (Figure 1a). The knobs are composed of two different tandem repeats, known as the 180 bp knob repeat and the 350 bp TR-1 repeat (Figure 1a). Also within the Ab10 haplotype are at least two trans-acting factors that independently convert the two types of knob into ‘neocentromeres.’ Moving rapidly along spindle microtubules, neocentromeres cause knobbed chromatids to segregate to the upper and lower cells of the linear tetrad (Dawe et al., 1999; Hiatt and Dawe, 2003a). As the lower cell of the tetrad develops into the egg, this mechanism gives Ab10 (and other knobs when Ab10 is present) a segregation advantage (Rhoades, 1952). In theory, Ab10 can reach 83% transmission as a heterozygote (Buckler et al., 1999), although measured meiotic drive levels typically range from 65–80% (Rhoades and Dempsey, 1966; Hiatt and Dawe, 2003a). The degree of preferential transmission is limited by a number of factors including recombination between centromeres and knobs and the efficiency of neocentromere formation (Hiatt and Dawe, 2003a).
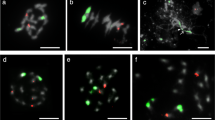
Structural variants of maize chromosome 10. (a) Schematic of the distal portion of the long arm of normal 10 (N10), abnormal 10 type 1 (Ab10-I), type 2 (Ab10-II) and type 3 (Ab10-III). N10 possesses no knobs and the Ab10s contain different amounts of TR-1 repeat (red) and knob 180 repeat (green). The subdomains of Ab10 are the TR-1 region, shared region, 180 bp knob and distal tip. There is a major inversion in the shared region that includes the W2, O7, L13 and Sr2 loci (shown in gray for Ab10-III, because it is assumed but has not been verified). (b) Chromosome 10 karyotypes. From left to right, FISH images from root tip spreads show N10, Ab10-I, Ab10-II, and Ab10-III. Cartoon depictions are shown below. A pachytene image from a male meiocyte shows the Ab10-III haplotype in greater detail. Images show DNA (gray), the centromere repeat CentC (yellow), TR-1 (red) and knob180 (green).
Although knobs are common in maize and teosinte (Albert et al., 2010), early cytological surveys suggest that Ab10 is relatively rare, segregating in only 18% of sampled maize landrace populations and 35% of teosinte populations (McClintock et al., 1981). Meiotic drive systems are often maintained at low levels in nature due to intrinsic deleterious fitness effects and/or selection for host-encoded suppressors that reduce meiotic drive (Lyttle, 1991; Burt and Trivers, 2006). Plausible explanations for the low frequency of Ab10 include a reduced male transmission of Ab10 or other fitness consequences of Ab10 in the homozygous condition (Buckler et al., 1999). In addition, there is evidence that the diversity and frequency of Ab10 has been affected by maize domestication. For example, the Ab10-I type originally described by Rhoades (1942 is the only known form of Ab10 in landraces (McClintock et al., 1981), while at least two forms are known in teosinte—the Ab10-I type and a cytologically distinct form known as Ab10-II (Kato, 1976; McClintock et al., 1981; Rhoades and Dempsey, 1985). Neither Ab10-I nor Ab10-II has been observed in any modern maize inbred lines (Albert et al., 2010).
In this study, we developed molecular markers for the Ab10 haplotype and used them to study the abundance and diversity of Ab10 in maize and its wild relatives. Prior data suggest that much of the Ab10 haplotype (both Ab10-I and Ab10-II) is derived from distal sections of the normal chromosome 10 (N10), but that the N10 sequences are scattered, rearranged and mixed with unknown sequence and transposable elements (Figure 1a) (Rhoades and Dempsey, 1985; Mroczek et al., 2006). Due to these rearrangements the Ab10 haplotype does not recombine with its N10 counterpart, although Ab10-I and Ab10-II can recombine with each other (Rhoades and Dempsey, 1985). As Ab10 rarely recombines with N10 but is nevertheless similar in sequence, we reasoned that if we sequenced long sections of the N10-Ab10 shared region from BAC clones we could identify sequences and/or polymorphisms unique to Ab10. This approach allowed us to identify two Ab10-specific PCR markers. In conjunction with an extensive new cytological survey, we use these markers to document the frequency of Ab10 in maize and related wild taxa. We find that Ab10 is more common in teosintes than cultivated maize landraces, and presumably absent altogether from modern inbreds. We document a previously unknown variant of Ab10 (Ab10-III) that shows a novel knob structure and appears to be more prevalent than Ab10-I or Ab10-II. Finally, an analysis of allele frequency differentiation of Ab10 across populations suggests that the factors that determine Ab10 abundance in natural and cultivated populations are complex.
Materials and methods
BAC library creation and gridding onto high-density filters
Tissue from a lab stock homozygous for the Ab10-I haplotype was used to create a bacterial artificial chromosome (BAC) library (ZMMTBa). This line has an undefined genetic background. The BAC library was created in the pIndigoBAC-5 vector as described previously (Peterson et al., 2000) using the HindIII restriction enzyme option and the ‘Y’ method for nuclei extraction with minor modifications (Peterson et al., 2002). BACs were individually archived in 384-well microtiter plates. Clones were gridded and fixed onto five nylon membranes as described previously (Magbanua et al., 2011). The average insert size of the clones was 112 kb, and the library afforded 4x coverage of the maize genome.
Overgo probe design and library hybridization
Single-copy cDNA sequences from normal chromosome 10L (B73 reference genome version 1) found between the marker rps11 (distal to the R1 locus) and the end of the chromosome were identified using BLAST at PlantGDB.org. Twenty-eight single-copy overgo probes were designed from the cDNA sequences and used for BAC library screening (Supplementary Table 1). Overgo probes were diluted to a working concentration of 20 μM, labeled with 32P-dCTP and 32P-dATP, pooled and hybridized to membranes at 55 °C (Magbanua et al., 2011). Of these 28, four did not hybridize to the library and 24 identified more than one BAC clone (between two and eleven colonies). In addition, several BACs hybridized to more than one probe (Supplementary Table 1). Ninety-six positive clones were handpicked, re-grown and spotted onto nylon membranes as a sub-library. The 96 colony membranes were re-probed with overlapping pools of probes to identify correspondence between colonies and probes. Eleven BACs were chosen for sequencing, and are referred to by the probe number(s) that hybridized to them in the sub-library screen. The original library coordinates for these 11 BACs are listed in Supplementary Table 2.
BAC preparation and sequencing
The 11 BACs were purified using the Large-Construct Kit (Qiagen, Valencia, CA, USA). Insert size was assayed by pulsed field gel electrophoresis of BACs digested with NotI (Peterson et al., 2002). Intact BACs were submitted to the Georgia Genomics Facility for 454 Titanium FLX sequencing. BAC sequences were submitted to the National Center for Biotechnology Information (NCBI) as high-throughput genome sequence phase 1 (HTGS-1) (see Supplementary Table 1 for GenBank numbers).
RNA isolation and cDNA preparation
Immature tassels were dissected from sibling plants containing Ab10 or the canonical chromosome 10 (N10). Anthers spanning pre-meiotic to mature pollen stages were frozen in liquid nitrogen and RNA was extracted using the Qiaquick RNA extraction kit (Qiagen). cDNA was synthesized using the Mint kit (Evrogen, Moscow, Russia), then normalized with the Trimmer kit (Evrogen). The Ab10 and N10 normalized cDNA libraries were submitted to Emory University for 100 bp paired-end Illumina sequencing. Each library was run on its own lane and reads were assembled into de novo cDNA contigs by the core facility at Emory. This resulted in the identification of 67 725 cDNA contigs from Ab10-I line, and and 46 498 contigs from the N10 line.
BAC sequence assembly and analysis
The 454 reads were assembled with both Newbler (454 Life Sciences, Roche, Branford, CT 06405, USA) and MIRA (http://sourceforge.net/apps/mediawiki/mira-assembler/). These assemblies and remaining raw reads were then further assembled using Sequencher (Gene Codes Corporation, Ann Arbor, MI, USA). Two BACs that hybridized to the same two probes (11–12) were assembled together and treated as a single BAC. To identify putative genes, repetitive sequences were first removed using RepeatMasker (http://www.repeatmasker.org/). Masked contigs were then mapped with BLAST to the non-redundant nucleotide and protein databases at NCBI with an e-value cutoff of 10−5 (Supplementary Table 2). Gene models were identified using FGenesH (SOFTBERRY) and Augustus (http://bioinf.uni-greifswald.de/augustus/).
Mapping BACs to Ab10 haplotypes
Nine repeat junction primers (RJ) were identified using RJPrimers: v1.0 from the unmasked BAC sequence contigs (Luce et al., 2006; You et al., 2010) (Supplementary Table 3). Six intron size polymorphism primers were identified by comparing the 35 complete genes identified in Ab10 BACs to their homologs in the B73 reference genome (Schnable et al., 2009) (Supplementary Table 3). Introns that differed in size by at least 50 bp between Ab10 and B73 were tested as markers. PCR was used to map the BACs within the Ab10 haplotype using the combined set of 15 primers in a series of DNAs extracted from deficiency lines (Hiatt and Dawe, 2003b). PCR was performed using 10–30 ng genomic DNA in reactions containing 1x Sigma-Aldrich (St Louis, MO, USA) PCR buffer, 2.5 mM MgCl2, 0.25 mM dNTPs, 0.25 μM primers and 1–2 unit Sigma Taq polymerase. Reactions were denatured at 94 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 55–59.5 °C for 20 s, 72 °C for 30–60 s and a final extension at 72 °C for 5 min.
Stocks used were Ab10-I_Rhoades (referred to as Ab10-I throughout), Ab10-II_Rhoades (referred to as Ab10-II throughout) and the deficiency lines Ab10-I-Df(I), Ab10-I-Df(F), Ab10-I-Df(H) and Ab10-I-Df(K), all originally obtained from Marcus Rhoades. Additional deficiency lines Ab10-I-Df(B), Ab10-I-Df(M) and Ab10-I-Df(L) were described previously (Hiatt and Dawe, 2003b), and Ab10-II-Df(Q) and Ab10-II-Df(M) were obtained from the Maize Genetics Cooperation Stock Center, University of Illinois, Urbana, Il. The deficiency lines were crossed to either the B73 or W23 (N10) inbreds, and the resulting heterozygotes used for scoring the markers by presence or absence.
Teosinte and landrace PCR screen
The two dominant RJ markers D6 (Ab10-D6) and G8 (Ab10-G8) (Supplementary Table 3) were used to screen a set of 638 DNA samples derived from 135 landraces, 10 populations of the lowland teosinte taxon Zea mays ssp. parviglumis and 10 populations of the highland teosinte taxon Zea mays ssp. mexicana (Supplementary Table 4). These two teosintes represent the closest wild relatives of maize (Matsuoka et al., 2002). Samples were selected to be both geographically and genetically representative based on previously published data (Fukunaga et al., 2005; Vigouroux et al., 2008; van Heerwaarden et al., 2011). Genomic DNA was prepared using a CTAB method (Saghaimaroof et al., 1984; Clarke, 2009). An additional 117 individuals from some of these 135 races, as well as 5 additional landraces were only screened with Fluorescence in situ hybridization (FISH) (see below). Thus, in total 769 individuals from 20 teosinte populations and 140 landrace populations were screened with FISH and/or PCR (Supplementary Tables 4, 5).
Fluorescence in situ hybridization
Mitotic root tip chromosomes from 267 landrace individuals were analyzed by FISH as described previously (Kato et al., 2004). Mixtures of oligo probes were used to visualize the 156 bp centromeric repeat, CentC and the two knob repeats, TR-1 (350 bp) and knob180 (180 bp). Fluorescently-labeled DNA oligos for TR-1 (20–21 nt long, four oligos) and CentC (20–23 nt long, four oligos) were obtained from Integrated DNA Technologies (http://www.idtdna.com/). The TR-1 oligos were 5′-Cy3 labeled and the CentC oligos were 5′ Cy5 labeled. Ten FITC-labeled DNA oligo probes for knob180 were previously designed (Yu et al., 1997). Each oligo was resuspended in 2x SSC (saline sodium citrate) buffer to 100 μM. The oligos for each repeat were mixed in equimolar amounts and diluted to a final concentration of 10 μM. The 10 μM probe mixtures were used directly for FISH of prepared slides. A solution of 0.5 μl TR-1 probe mix, 0.5 μl CentC probe mix, 0.2 μl knob180 probe mix, 5 μl salmon sperm DNA (140 ngμl−1) and 3.8 μl 2x SSC in 1x TE (TRIS EDTA) buffer was dropped onto slides containing the chromosomes. Slides were then denatured 5–10 min in a humid chamber in a boiling water bath and allowed to hybridize at room temperature for 1–3 h. Slides were then rinsed in 2x SSC, air-dried and mounted in 10 μl of Vectashield with DAPI (Vector Laboratories, Burlingame, CA, USA). Meiotic chromosomes were prepared as previously described (Shi and Dawe, 2006) and subjected to the same FISH protocol. Images were collected using a Zeiss Axio Imager and processed using Slidebook 5.0 software (Intelligent Imaging Innovations, Denver, CO, USA).
Estimating the divergence of the Ab10 haplotypes
Two regions of the Ab10 haplotype (Figure 2) were amplified and PCR products were directly sequenced from both directions from 11 different chromosomes (seven N10 and four Ab10). The two loci assayed (3471 and 8042) can be found on chromosome 10 in the B73 genome (version 2) at positions 142093638–142094095 (3471=GRMZM2G119802) and 147609512–147609977 (8042=GRMZM2G150286). These regions, 348 bp from locus 3741 and 467 bp from locus 8042, include portions of exonic and intronic sequences. For N10 controls, we used lines from the USDA Germplasm Resources Information Network (GRIN) (http://www.ars-grin.gov/npgs): Mo17 (PI 648432), CML220 (Ames 27087), B73 (PI 550473), I137TN (Ames 27116), Tzi9 (PI 506247), K55 (Ames 22754) and CI66 (PI 587148). Two isolates of Ab10-I were used, Ab10-I_Rhoades, originally collected from a site outside of Mexico city, and a second isolate (line X233F from the Maize Genetics Cooperation Stock Center) that was collected from the American Southwest. Our Ab10-II stock was obtained from Marcus Rhoades. The Ab10-III chromosome was identified in a homozygous state in the landrace Guatemala 110 (PI 490825 from the USDA Germplasm Resources Information Network).
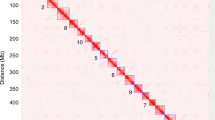
Positions of probes on N10 and corresponding BACs on Ab10-I and Ab10-II. The relative positions of 28 probes used to probe the Ab10 BAC library are shown on N10. The positions of known gene markers (rsp11, gln1 and so on), the loci used for molecular dating (3741 and 8042, blue), and the estimated position of the two dominant markers D6 and G8 (purple) are also shown. The mapped locations of the Ab10 BACs are shown between deficiency breakpoints ((Df(B), Df(C) and so on). The general maps of Ab10-I and Ab10-II (including marker positions) are adapted from previous work (Mroczek et al., 2006), with TR-1 shown in red and knob 180 shown in green.
The sequences were used to estimate phylogeny and haplotype divergence between Ab10 and N10. In order to investigate the phylogenetic relationship of N10 and Ab10 we aligned sequences in MEGA (Tamura et al., 2007) and created maximum likelihood trees with 500 bootstrap iterations, using a Jukes–Cantor nucleotide substitution model. The divergence time between haplotype groups was estimated as T=2 Lμ/d, where T is the divergence time in years, L the length of the sequence in bp, μ the mutation rate per bp and d the distance between haplotypes estimated using counts of fixed differences or Nei’s net divergence (Nei, 1987). In all cases, we assumed a mutation rate of 3 × 10−8, similar to recent estimates from maize (Clark et al., 2005).
Frequency of Ab10 in teosinte populations
We used the software BayeScan 2.01 (Foll and Gaggiotti, 2008) to investigate whether Ab10 frequencies in teosinte populations deviate from neutral expectations. The software models allele frequencies by estimating population effects common to all loci and locus-specific effects common to all populations. If the effect of an individual locus is estimated to be different from zero, selection is inferred. The posterior probability of a nonzero locus-specific effect is evaluated by comparing the probabilities of models with and without selection using a Markov Chain Monte Carlo approach. We conducted the analysis for both subspecies together and separately to account for the hierarchical population structure of the two teosinte subspecies (Pyhäjärvi et al., 2012). SNPs genotyped in the same teosinte individuals (Pyhäjärvi et al., 2012) used in our Ab10 PCR screen were transformed into dominant markers by randomly treating one allele as recessive. These SNPs exhibit unusually high average expected heterozygosity (HE) because of the ascertainment scheme of the markers included on the genotyping chip. To remove this effect, only 1907 SNPs that had similar HE (0.05–0.1) to the D6 and G8 Ab10 PCR markers (HE 0.09 and 0.07) were included in the analysis. Estimated heterozygosities were based on the assumption of Hardy–Weinberg equilibrium. BayesScan was run with default parameters: 20 pilot runs, a burn-in period of 50 000 iterations and 5000 output iterations with a thinning interval of 10. Prior boundaries for the inbreeding coeffcient (FIS) for each population were from 0 to 0.1 based on Pyhäjärvi et al. (2012).
Results and discussion
Sequence of Ab10 BACs
We created a BAC library from a line homozygous for Ab10-I and screened it with multiple probes distributed across the region shared between N10 and Ab10 (Supplementary Table 1, Figure 2). Eleven BACs were sequenced using 454 technology (two of which overlapped and were treated as one). The sequences were assembled into unordered contigs and processed using gene prediction programs. By this assay at least 43 genes were observed (Supplementary Table 2). We also sequenced the transcriptomes from a single Ab10-I plant and a wild-type (N10) sibling and confirmed that at least 35 of the Ab10-I genes were transcribed (see Materials and Methods and Supplementary Table 2). All but one of the 35 confirmed Ab10 genes showed clear homology to known genes on the long arm of normal chromosome 10 in the B73 reference genome. In the single exceptional case, the homolog in B73 is present on chromosome 9 (GRMZM5G811697).
Development of molecular markers to map Ab10 BACs
One defining feature of Ab10-I and Ab10-II is that they do not recombine with N10 distal to the R1 locus, making traditional mapping a challenge (Rhoades, 1942). Deletion mapping is a viable alternative as large terminal deficiencies of Ab10 can be transmitted through the female (Hiatt and Dawe, 2003b). Previous studies have used deficiencies of Ab10-I and Ab10-II to map shared genes with mutant phenotypes: White Seedling2 (W2), Opaque Endosperm7 (O7), Luteus13 (L13) and Striate Leaves2 (Sr2) (Rhoades and Dempsey, 1985; Rhoades and Dempsey, 1988). Ab10-I deficiencies have also been used to map RFLP markers (Mroczek et al., 2006). In principle, the Ab10-I and Ab10-II deficiencies can be used to map any marker, as long as the marker is dominant or co-dominant, as nearly all of the Ab10 deficiencies are homozygous inviable and must be propagated as heterozygotes with N10 (Hiatt and Dawe, 2003b).
We identified at least one PCR marker per BAC (Supplementary Table 3). Among these were six intron size polymorphisms and nine RJ markers (Luce et al., 2006; You et al., 2010) that we used to differentiate Ab10 from N10 in the genetic backgrounds of our heterozygous Ab10 deletion lines. For these tests, the N10 chromosomes were derived from either the W23 or B73 inbreds. Eight deletion lines exist for Ab10-I, however, only two deletion lines exist for Ab10-II, and these two have been poorly characterized (Rhoades and Dempsey, 1988). By scoring the presence or absence of the markers in the Ab10 deletion lines we established that BAC2-3 mapped upstream of the most proximal known breakpoints of both Ab10-I and Ab10-II (Figure 2). On Ab10-I, all other mapped BACs are located between the Df(I) and Df(F) breakpoints, though the sequences of these BACs do not overlap. This finding is consistent with prior data confirming the existence of a large inversion within the Ab10 haplotype. A similar inversion exists on Ab10-II. Four BACs that correspond to the proximal side of Ab10-II (between Df(Q) and Df(M)) map to the distal side of N10, and four others from the distal region of Ab10-II map to the proximal side of N10 (Figure 2). None of the BACs, which span nearly all of the shared region on N10, map distal to the Ab10-I-Df(L) breakpoint. These data show that the strongest homology between N10 and Ab10 lies in the regions proximal to the major knob. The ‘distal tip’ is not derived from modern chromosome 10 and its origin remains obscure, yet it is known to contain at least one of the key meiotic drive functions (Hiatt and Dawe, 2003a).
Identification of Ab10-specific markers for population screens
The dominant molecular markers we used to map Ab10 BACs can also be used in a diagnostic fashion to identify the chromosome in cultivated or natural populations of Zea. However, we needed to first confirm that they were unique to Ab10. Prior cytological data had established that Ab10 is not present in at least 103 traditional maize inbreds (Albert et al., 2010). We tested our 15 Ab10-based primer pairs in a similar set of 53 inbreds (Shi et al., 2010) that includes 24 lines not assayed in Albert et al. (2010). Thirteen of the markers were not consistently polymorphic between Ab10 and N10, and were not used in population screens. However, two of the dominant RJ PCR markers, Ab10-D6 (‘D6’) and Ab10-G8 (‘G8’), were not found in any of the 53 inbred lines, suggesting they are unique to Ab10. To further test their reliability, we carried out a larger test screen that combined PCR with FISH on a total of 150 individuals from 19 maize landraces (Supplementary Tables 4, 5). A total of 34 individuals contained Ab10 as scored by both PCR and FISH. The PCR and FISH results were entirely concordant, although many Ab10 chromosomes carried only one of the two markers (either D6 or G8) (Supplementary Tables 4, 5). Another 116 individuals tested negative for Ab10 by both FISH and PCR (Supplementary Tables 4, 5). The observation that only two of our 15 Ab10-derived markers were unique to Ab10 supports the general conclusion that Ab10 is very similar to N10 in the shared region. The fact that the two markers do not always co-occur suggests that there is sequence-level variation between cytologically similar Ab10 haplotypes.
Discovery of a new cytological variant, Ab10-III
While performing FISH on our landraces, we observed that four landraces were segregating Ab10-I, and unexpectedly, that nine landraces carried a new cytological variation of the Ab10 haplotype that we term Ab10-III. Ab10-III appears to be structurally similar to Ab10-I in the central domain, but contains a second TR-1-rich domain appended to the end of the main knob (Figure 1b). We do not yet know if Ab10-III exhibits neocentromere activity and meiotic drive, but it seems likely based on the strong structural similarity to the other Ab10 types. Moreover, as the two major knob repeats have independent neocentromere activities, with TR-1 moving poleward faster than the knob180 repeat (Hiatt et al., 2002), we speculate that the additional TR-1 knob found on Ab10-III may provide this haplotype with an advantage over other Ab10 haplotypes.
Notably, the two dominant markers D6 and G8 cannot be used to differentiate the different Ab10 haplotypes and do not consistently co-occur with major cytological features (Supplementary Tables 4, 5). Although the Ab10-I_Rhoades haplotype used to create the BAC library carries both markers, three other landraces with chromosomes that appear cytologically similar to Ab10-I scored positive for G8 but not D6 (Supplementary Table 5). Ab10-III was observed to have D6, G8, or both D6 and G8 depending on the isolate (Supplementary Table 5). This finding supports the view that Ab10 haplotypes recombine with each other even though they rarely if ever recombine with N10 (Rhoades and Dempsey, 1985).
Estimating divergence time of haplotypes
The relationship of Ab10 and N10 was examined by sequencing two loci in the shared region, one from BAC 2/3 (3741) and another from BAC 16 (8042) (Figure 2). Gene trees at both loci revealed an unexpected grouping pattern (Figure 3, Supplementary Figure 1), in which Ab10-I and Ab10-II form one group while Ab10-III consistently groups with N10 haplotypes. These data reveal that while Ab10-I and Ab10-II are clearly related, Ab10-III appears to have either arisen independently on an N10 background or exchanged material with N10 haplotypes via gene conversion. The Ab10 (-I and -II) and N10 (+Ab10-III) groups differ at 11 fixed SNPs within the 348 bp locus 3741 and 8 SNPs within the 467 bp locus 8042. Estimates of divergence time based on these fixed SNPs suggest the groups split ∼365 000 years ago. Estimates based on net pairwise nucleotide divergence (Nei, 1987), suggest the split occurred 535 000 years ago for locus 3741 and 377 000 years ago for locus 8042. These results do not differ appreciably from similar estimates of many other maize genes (Doebley and Iltis, 1980; Ross-Ibarra et al., 2009), and suggest that the shared region either arose fairly recently in the maize lineage, or has sustained a low level of gene conversion with N10 chromosomes.
Maximum likelihood phylogeny of locus 3741 for N10 inbreds and homozygous Ab10 stocks. K10L2 is an N10 variant described by McClintock et al. (1981) as having a knob on its long arm.
Ab10 abundance and allele frequencies in teosinte and landraces
Using the D6 and G8 markers, as well as FISH assays, we scored a total of 769 Zea mays (maize and teosinte) individuals for Ab10. The D6 and G8 markers were used to assess the frequency of Ab10 in 10 natural populations of Zea mays ssp. parviglumis, and 10 natural populations of Zea mays ssp. mexicana at a sample size of 12 individuals each (for a total of 240 individuals) (Supplementary Table 4). In addition, 529 individuals from 140 landraces were scored using PCR and/or FISH (including the 150 landrace individuals used in our Ab10-specific marker screen above; Supplementary Tables 4, 5). The results are summarized in Figures 4 and 5. The Figure 5 also compares our Ab10 frequency data to prior data accumulated by McClintock et al., 1981. The same trends are apparent in both data sets: Ab10 is more common in teosinte populations than maize landrace populations, and Ab10 is rarely observed in >40% of the individuals in any given population.
Map of screened populations showing Ab10 frequency for each population. Locations of landrace (blue), mexicana (red) and parviglumis (green) populations in North and South America (a). Zoomed in views of Mexico (b) and South America (c) are shown. The frequency of Ab10 is shown as the shaded area in each pie chart.
Ab10 frequency comparison of current study and study by McClintock et al. (1981). Values shown are percentage of positives out of total sampled. Teosinte subspecies designations were not known at the time of the McClintock et al. (1981) study.
These data revealed that Ab10 is found at an average frequency of 14% in parviglumis (17 positive individuals from 7 populations), 16% of mexicana (19 positive individuals from 8 populations), and 13% of maize landraces (68 positive individuals). The two markers differ in frequency among the three subspecies: G8 is more prominent in landraces and parviglumis compared with D6, but D6 is more common in mexicana (Supplementary Tables 4, 5). Among the landrace individuals scored by FISH, we observed Ab10-I infrequently, never observed Ab10-II and observed Ab10-III most frequently (Supplementary Tables 4, 5). Previous work suggests that Ab10-II is a common variant in teosintes (Kato, 1976; McClintock et al., 1981), but we were unable to verify this via FISH assays.
Overall our data demonstrate that Ab10 is more common and more diverse than originally thought. At frequencies of 13–16%, different variants of Ab10 must regularly come into contact with each other, where they presumably recombine in the proximal regions, and compete with each other for preferential transmission. As knobs on other chromosomes compete for transmission based on their size (Kikudome, 1959), it follows that head-to-head competition between different Ab10 variants will favor haplotypes with larger knobs. This could explain why Ab10-III (which has the most knobs) is the most prevalent haplotype in maize.
Population differentation of Ab10
Although the frequency of Ab10 varied among populations, we found little evidence for unusual patterns of allele frequency differentiation. Comparison of the posterior probability of a locus-specific effect (interpreted as evidence of selection) for markers D6 and G8 with probabilities for nearly 2000 randomly chosen SNPs revealed no support for selection on either marker (Figure 6a), especially compared with the subset of SNPs previously shown to correlate with environmental variables ((Pyhäjärvi et al., 2012). Although Ab10 frequency shows a negative correlation with elevation in both teosinte subspecies (Figure 6b, r=−0.45 for each) and landrace maize (Figure 5b; P=0.027, r=−0.39), only the latter is statistically significant.
Population structure and clinal variation at Ab10. (a) BayesScan analysis of FST for PCR markers G8 and D6 in teosinte populations. Shown are the distributions of FST (y axis) and the log10 ratio of the posterior probability of a model including selection (x axis). Higher values of the log posterior odds indicate stronger evidence for selection. Data are from 1907 random SNPs (Pyhäjärvi et al., 2012) (transparent black) and Ab10 (red). For comparison, green points show SNPs strongly associated (top 1%) with environmental variables Pyhäjärvi et al., 2012. The analysis was done separately for parviglumis, mexicana and for the two combined. Vertical lines indicate a false discovery rate threshold of 5%. The horizontal gray bar indicates 5–95% quantiles of FST. (b) Regression of Ab10 frequency against elevation for each subspecies. Data for maize are based on accessions pooled into 100 m altitudinal bins.
Meiotic drive alone should cause Ab10 to quickly fix across populations (Buckler et al., 1999). The observation that Ab10 is not fixed in any population suggests that additional, intrinsic (homozygous disadvantage, pollen inviability and so on) and/or extrinsic (environmental effects, gene flow, population colonization history) factors are limiting Ab10 frequency. Ab10 does have negative effects on pollen viability (Rhoades, 1942) and may have negative fitness effects when homozygous, but this has not been tested. A combination of uniform meiotic drive and consistent negative selection should lead to an equilibrium frequency of Ab10 found across all populations. However, we observed no evidence for significantly lower differentiation of our Ab10-specific markers, suggesting that environmental factors also have a significant impact Ab10 frequency among populations. Clinal patterns of genome size in maize imply selection against large genomes at high elevation (Poggio et al., 1998). As chromosomal knobs are the major determinant of genome size variation in maize and teosinte (Chia et al., 2012), selection against genome size may explain the observed negative correlation between Ab10 and elevation. We cannot rule out other possibilities, however, including population size and selection efficiency or direct environmental differences in the fitness of Ab10 (Buckler et al., 1999).
Data archiving
BAC sequence data have been submitted to GenBank: JX646739- JX646748.
References
Albert PS, Gao Z, Danilova TV, Birchler JA (2010). Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet. Genome Res. 129: 6–16.
Buckler ES, Phelps-Durr TL, Buckler CSK, Dawe RK, Doebley JF, Holtsford TP (1999). Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426.
Burt A, Trivers R (2006) Genes in conflict: the biology of selfish genetic elements. Harvard University Press: Cambridge, MA, USA.
Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J et al (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet 44: 803–807.
Clark RM, Tavare S, Doebley J (2005). Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus. Mol Biol Evol 22: 2304–2312.
Clarke JD (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harbor Protocols 4: 5177.
Dawe RK, Reed LM, Yu HG, Muszynski MG, Hiatt EN (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11: 1227–1238.
Doebley JF, Iltis HH (1980). Taxonomy of Zea (Gramineae). 1. A subgeneric classification with key to taxa. Am J Bot 67: 982–993.
Foll M, Gaggiotti O (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180: 977–993.
Fukunaga K, Hill J, Vigouroux Y, Matsuoka Y, Sanchez J, Liu KJ et al (2005). Genetic diversity and population structure of teosinte. Genetics 169: 2241–2254.
Hiatt EN, Dawe RK (2003a). Four loci on abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164: 699–709.
Hiatt EN, Dawe RK (2003b). The meiotic drive system on maize abnormal chromosome 10 contains few essential genes. Genetica 117: 67–76.
Hiatt EN, Kentner EK, Dawe RK (2002). Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell 14: 407–420.
Kato A, Lamb J, Birchler J (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101: 13554–13559.
Kato YTA (1976). Cytological studies of maize (Zea mays L.) and teosinte (Zea mexicana Shrader Kuntze) in relation to thier orIgin and evolution. Mass Agric Exp Sta Bull 635: 1–185.
Kikudome G (1959). Studies on the phenomenon of preferential segregation in maize. Genetics 44: 815–831.
Luce AC, Sharma A, Mollere OSB, Wolfgruber TK, Nagaki K, Jiang JM et al (2006). Precise centromere mapping using a combination of repeat junction markers and chromatin immunoprecipitation-polymerase chain reaction. Genetics 174: 1057–1061.
Lyttle TW (1991). Segregation distortors. Ann Rev Genet 25: 511–557.
Magbanua ZV, Ozkan S, Bartlett BD, Chouvarine P, Saski CA, Liston A et al (2011). Adventures in the Enormous: a 1.8 Million Clone BAC Library for the 21.7 Gb Genome of Loblolly Pine. Plos One 6: e16214.
Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99: 6080–6084.
McClintock B, Yamakake T, Blumenschein A (1981) Chromosome Constitution of Races of Maize: its significance in the interpretation of relationships between races and varieties in the Americas. Colegio de Postgraduados: Chapingo, Mexico.
Mroczek RJ, Melo JR, Luce AC, Hiatt EN, Dawe RK (2006). The maize Ab 10 meiotic drive system maps to supernumerary sequences in a large complex haplotype. Genetics 174: 145–154.
Nei M (1987) Molecular evolutionary genetics. Columbia University Press: New York, NY, USA.
Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH (2000). Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide. J Agric Genomics 5: 1–100.
Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH (2002) Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide 2nd edn pp 1–91. http://www.mgel.msstate.edu/pubs/bacman2.pdf.
Poggio L, Rosato M, Chiavarino AM, Naranjo CA (1998). Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Annals Bot 82: 107–115.
Pyhäjärvi T, Hufford MB, Mezmouk S, Ross-Ibarra J (2012) Complex patterns of local adaptation in teosinte. arXiv:12080634v1 http://arxiv.org/abs/1208.0634.
Rhoades M (1942). Preferential segregation in maize. Genetics 27: 395–407.
Rhoades M (1952). Preferential segregation in maize. In: Gowen. JW, (eds) Heterosis Iowa State College Press Iowa State College Press: Ames, IA, USA. pp 66–80.
Rhoades M, Dempsey E (1985). Structural heterogeneity of chromosome 10 in races of maize and teosinte. In: Freeling M, Alan R (eds) Plant Genetics. Liss: New York, NY, USA. pp 1–18.
Rhoades M, Dempsey E (1988). Structure of K10-II chromosome and comparison with K10-I. maize coop. Newsletter 62: 33–34.
Rhoades MM, Dempsey E (1966). Effect of Abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics 53: 989–1020.
Ross-Ibarra J, Tenaillon M, Gaut BS (2009). Historical divergence and gene flow in the genus Zea. Genetics 181: 1397–1409.
Saghaimaroof MA, Soliman KM, Jorgensen RA, Allard RW (1984). Ribosomal DNA spacer-length polymorphisms in barley - Mendelian inheritance, chromosomal location, and popyulation dynamics. Proc Natl Acad Sci USA 81: 8014–8018.
Sandler L, Novitski E (1957). Meiotic drive as and evoluationary force. Am Nat 91: 105–110.
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S et al (2009). The B73 Maize Genome: complexity, diversity, and dynamics. Science 326: 1112–1115.
Shi J, Dawe RK (2006). Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583.
Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK (2010). Widespread gene conversion in centromere cores. PLoS Biol 8: e1000327.
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24: 1596–1599.
van Heerwaarden J, Doebley J, Briggs WH, Glaubitz JC, Goodman MM, JdJ SanchezGonzalez et al (2011). Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA 108: 1088–1092.
Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Jesus Sanchez G, Doebley J (2008). Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Am J Bot 95: 1240–1253.
You FM, Wanjugi H, Huo N, Lazo GR, Luo M-C, Anderson OD et al (2010). RJPrimers: unique transposable element insertion junction discovery and PCR primer design for marker development. Nucleic Acids Res 38: W313–W320.
Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK (1997). Neocentromere-mediated chromosome movement in maize. J Cell Biol. 139: 831–840.
Acknowledgements
We would like to thank Lauren Sagara for help with DNA extractions. Xueyan Shan, Melanie Smith, Calla Kingery and Zenaida Magbanua provided help with BAC library construction and probing. We are grateful to Ryan Weil for helping with cDNA sequence assembly, Saravanaraj Ayyampalayam for helping with BAC sequence assembly, and Michael McKain for general bioinformatics guidance. This work was supported by a grant from the National Science Foundation (NSF-0951091).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Kanizay, L., Pyhäjärvi, T., Lowry, E. et al. Diversity and abundance of the abnormal chromosome 10 meiotic drive complex in Zea mays. Heredity 110, 570–577 (2013). https://doi.org/10.1038/hdy.2013.2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2013.2
Keywords
This article is cited by
-
The maize abnormal chromosome 10 meiotic drive haplotype: a review
Chromosome Research (2022)
-
Unexpected patterns of segregation distortion at a selfish supergene in the fire ant Solenopsis invicta
BMC Genetics (2018)