Abstract
Estimates of genetic diversity represent a valuable resource for biodiversity assessments and are increasingly used to guide conservation and management programs. The most commonly reported estimates of DNA sequence diversity in animal populations are haplotype diversity (h) and nucleotide diversity (π) for the mitochondrial gene cytochrome c oxidase subunit I (cox1). However, several issues relevant to the comparison of h and π within and between studies remain to be assessed. We used population-level cox1 data from peer-reviewed publications to quantify the extent to which data sets can be re-assembled, to provide a standardized summary of h and π estimates, to explore the relationship between these metrics and to assess their sensitivity to under-sampling. Only 19 out of 42 selected publications had archived data that could be unambiguously re-assembled; this comprised 127 population-level data sets (n⩾15) from 23 animal species. Estimates of h and π were calculated using a 456-base region of cox1 that was common to all the data sets (median h=0.70130, median π=0.00356). Non-linear regression methods and Bayesian information criterion analysis revealed that the most parsimonious model describing the relationship between the estimates of h and π was π=0.0081h2. Deviations from this model can be used to detect outliers due to biological processes or methodological issues. Subsampling analyses indicated that samples of n>5 were sufficient to discriminate extremes of high from low population-level cox1 diversity, but samples of n⩾25 are recommended for greater accuracy.
Similar content being viewed by others
Introduction
Intra-specific surveys of genetic diversity are increasingly being used to assist in the conservation and management of biodiversity. They involve the comparison of samples obtained from different locations or from a time-series at the same location, generally referred to as ‘population-level’ samples in literature. DNA sequence variation in these population-level surveys is often quantified using data from haploid loci, with the mitochondrial gene cytochrome c oxidase subunit I (cox1, COI) being the most frequently sequenced in animals. The popularity of cox1 stems from the availability of several sets of conserved PCR primers (for example, Folmer et al., 1994) and the dual purpose of cox1 as a molecular marker for both intra-specific variation and species identification (Bucklin et al., 2011).
Several metrics are used for assessing diversity in population-level cox1 sequencing surveys. These include the absolute number of haplotypes, the number of unique or ‘private’ haplotypes, the probability that two randomly chosen haplotypes are different (haplotype diversity, h; Nei, 1987) and the average number of nucleotide differences per site between two randomly chosen DNA sequences (nucleotide diversity, π; Nei and Li, 1979). Other reported metrics, such as Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997), are typically used for assessing demography and/or selection rather than diversity.
By far, the most commonly reported diversity metrics in population-level cox1-sequencing studies are h and π. These metrics are useful for biodiversity assessment because they can be influenced by a variety of factors such as the proportion of asexual to sexual reproduction, the size and age of populations, the degree of connectivity between populations, the extent of introgression from related species, the underlying mutation rate and the impact of selection (for example, Boyer et al., 2007; Cárdenas et al., 2009; Haig et al., 2010; see also Bazin et al., 2006; Wares, 2010). In addition to these biological factors, population-level estimates of cox1 h and π are also affected by methodological issues. The best known of these is under-sampling. A low number of sampled individuals can artificially inflate or deflate h and π estimates (Nei and Li, 1979; Nei, 1987). Other powerful sources of bias come from the inadvertent inclusion of cryptic species (for example, Knowlton, 1986) in ‘single species’ samples and through the PCR co-amplification of nuclear-mitochondrial sequences (numts; Lopez et al., 1994) together with the target mitochondrial locus cox1. These unwanted ‘DNA contaminants’ have the potential to increase h and substantially increase π estimates. It is also possible that DNA sequencing errors may have an impact on the estimation of population-level h and π. This may be negligible in many instances but could be a problem for samples with low molecular diversities owing to a lower signal-to-noise ratio (Clark and Whittam, 1992).
Even though population-level estimates of cox1 h and π are widely used, there are a number of fundamental issues associated with the comparison of these metrics that have yet to be addressed. One of these is the data set re-assembly. DNA sequence data set re-assembly is required in order to make accurate comparisons against published results and to reduce bias in meta-analyses. Re-assembled DNA sequence data sets provide these benefits because they can be edited to include only homologous positions and analyzed using exactly same methods. Data set re-assembly depends on the use of archived sequence data; however, not all of the sequence data used in published studies is archived. Some authors lodge complete population-level samples of DNA sequences in archives (for example, GenBank; www.ncbi.nlm.nih.gov/genbank/), others submit only those sequences that differ (unique haplotypes) to archives and provide frequency/population identifier tables in the corresponding publications, whereas other authors do not archive any of the sequences generated for their publications at all. There is currently no information available on how many of the cox1 population-level data sets used in publications can be re-assembled from archived sequence accessions.
It is difficult for researchers to readily assess the wider significance of population-level cox1 h and π estimates because there is no published summary of this data. Specific comparisons between h or π from different studies are rare and context-dependent, involving species sharing taxonomic/ecological characteristics (for example, Kim et al., 2009) or population-level samples with similar levels of diversity (for example, Goodall-Copestake et al., 2010). Furthermore, similar values of h or π may be referred to as low in one publication but high in another. Quantitative and qualitative comparisons of population-level cox1 h and π would benefit from a summary of published diversity estimates that is taxonomically broad and standardized to a homologous region of the cox1 gene.
Two other issues that warrant attention are the haplotype–nucleotide diversity relationship and the impact of under-sampling on these metrics. Bird et al. (2007) described a positive relationship between cox1-based population-level estimates of h and π in a study on three species of Hawaiian limpet (Cellana spp.). A greater understanding of the nature of this relationship may provide a useful aid to the interpretation of pairs of h and π estimates. However, the haplotype–nucleotide diversity relationship has not been assessed quantitatively or over a taxonomically broader range of samples. Likewise, little attention has been paid to the impact of sample size on the estimation of population-level h and π. Low sample numbers (for example, n⩽5) are common in intra-specific sequencing studies due to difficulties associated with sample collection and the high cost of first generation (Sanger) DNA sequencing. Although it is generally appreciated that low sample numbers can lead to bias, it is not clear how strong this bias may be for population-level estimates of cox1 h and π.
In this study, we begin to address these fundamental issues associated with the comparative analysis of population-level cox1 diversity data. Specifically, we aim to quantify how many data sets from cox1 population-level studies can be re-assembled and to use re-assembled data sets to provide a working series of standardized h and π estimates. We also aim to model the relationship between population-level estimates of cox1 h and π and to assess the sensitivity of these metrics to low sample sizes.
Materials and methods
Science literature databases were queried using the terms cytochrome c oxidase subunit I, cox1 and COI. The results of this search were manually examined for publications reporting estimates of h and π for samples of n⩾15 individuals from a single species per locality that were derived by analyzing ⩾450 bases from the 5′ region of the cox1 gene. The sample size of n⩾15 individuals was chosen as a compromise between the benefit of having a large comparative data set and the cost of bias introduced from small sample sizes. Criteria for species and sampling-site selection could not be refined because species and sampling-site boundaries differed between studies depending on the opinions of the corresponding authors. Only studies using overlapping fragments of cox1 DNA sequence were considered to ensure that subsequent comparative analysis could be performed using exactly the same (homologous) nucleotide sites within the gene. To maximize species coverage and fragment overlap, we focused on the 5′ end of cox1 as this is most commonly sequenced region of the gene and defined a minimum sequence length cutoff of 450 bases.
Population-level cox1 sequence data sets from studies that matched our selective criteria were manually re-assembled using GenBank accessions and associated haplotype frequency/population identifier data within the corresponding publications. Estimates of h and π were calculated from each data set using the software DNASP v.5.10.01 (Librado and Rozas, 2009) and the results compared against those reported in the publications. Data sets that generated values of h and π which differed from the published results were excluded from subsequent analysis, if these discrepancies could not be explained by the rounding of numbers or obvious (correctable) formatting errors. New diversity estimates for each of the remaining data sets were then calculated using DNASP from a homologous 456-base region of cox1 that was common to all of the sequences. This DNA region corresponds to positions 136–591 within the full-length sequence of many invertebrate cox1 genes, for example the bumblebee (Bombus ignitus) cox1 gene in the mitochondrial genome accession NC_010967.
The relationship between h and π was investigated using the newly derived estimates obtained from the re-assembled 456-base data sets. Different potential models evaluating the relationship between the two metrics were compared based on the functional forms of h and π using least-squared non-linear regression methods. Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) were used to examine relative model fit (Burnham and Anderson, 2002) using the software Mathematica v.8 (Wolfram Research, Long Hanborough, UK). Deviations from the fitted model were transformed as the square root of the absolute residuals. 75th and 95th deviation percentiles were then calculated from the transformed values.
To assess the effect of sample size on population-level estimates of cox1 h and π we used re-assembled data sets for the bumblebee Bombus ardens (Kim et al., 2009) and Antarctic krill Euphausia superba (Goodall-Copestake et al., 2010). These species were selected to represent low- and high-diversity scenarios, respectively. Data sets of n=50 individuals for each species were assembled by randomly selecting cox1 sequences from three location-specific samples of B. ardens (Hadandong, Jeongseon, Ulleungdo) among which there was no evidence for genetic structure (Kim et al., 2009) and from a single swarm-specific net sample (number 2) of E. superba (Goodall-Copestake et al., 2010). Sample order within both of the n=50 data sets was randomized 100 times and subsamples of 2–49 individuals were selected from each randomization. DNASP was used to calculate h and π from the 456-base region of cox1 (defined above) for the starting sample of n=50 and for each of the randomized subsamples.
Results
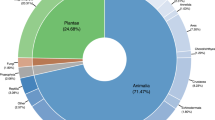
An extensive search of over 500 publications identified 42 studies that matched our selection criteria (estimates of both h and π, samples of n⩾15 individuals per location, ⩾450 bases sequenced from the 5′ end of cox1). Only 19 publications provided sufficient information to allow full re-assembly of population-level data sets (Figure 1). The main impediments to data set re-assembly were a lack of either archived DNA accessions (11 publications) or a lack of frequency data for archived unique haplotype lists (14 publications). Another factor was that some of the re-assembled data sets produced results that differed from those in the corresponding publications, even after taking into account discrepancies due to the rounding of numbers. Differences in six data sets could be corrected for, because they concerned obvious formatting errors within the publications (for example, the misplacement of a decimal place within a results table); however, 16 of the differences between re-assembled data sets and published results could not be resolved (Figure 1).
Functional breakdown of the data acquisition procedure. The upper pie chart represents the number of candidate population-level data sets found in publications that matched our search criteria and the lower pie chart the number of data sets re-assembled. Numbers in brackets after the data set descriptions follow: number of population-level data sets, number of corresponding publications.
Data from a total of 127 population-level samples (locations) and 23 species were unambiguously re-assembled, with representatives of Crustacea and Mollusca dominating (Table 1). The number of population-level samples per species ranged from 1 to 14 (mean=6, median=5) and the number of individuals per sample ranged from the minimum cutoff of 15 to 91 (mean=24, median=20). DNA sequence lengths ranged from 494 to 1173 bases (mean=666, median=650). No stop codons were inferred in amino acid translations. Reducing the original variable-length cox1 sequence data sets to a homologous 456-base region did not cause any of the population-level samples with h>0.00000, π>0.00000 to become h=0.00000, π=0.00000. Estimates of h and π derived from the 456-base sequences differed from estimates from the variable-length sequences on average by a factor of 0.08808 and a factor of 0.16373, respectively.
Haplotype diversity for the 456-base data sets ranged from 0.00000 to 0.98506 with a mean h=0.63388 that was slightly smaller than the median (h=0.70130). Nucleotide diversity ranged from 0.00000 to 0.01993 with a mean π=0.00388 that was slightly larger than the median (π=0.00356). Values for both of these cox1 diversity metrics encompassed a near-continuous range with the exception of one extreme π outlier corresponding to a single sample of the mite harvestman Rakaia denticulata (Table 1, Figures 2 and 3). There was a considerable amount of variation in both h and π estimates among population-level samples within species relative to the variance of h and π within the total data set (Table 1, Figure 2). Population-level samples of the shrimp Phycomenes zostericola (n=10) exhibited the greatest within species variation, with estimates of h covering 98% of the total data set range and π covering 49% of the total range. Only a few species had population-level samples with similar levels of diversity, for example E. superba for which the variance in both h and π of population-level samples (n=9) was <10% of the total data set range.
Variance in estimates of h and π among population-level samples within species (n=23) calculated from a homologous 456-base region of the cox1 gene. Median values for the total sample set (n=127) are shown as dashed vertical lines. Species order follows the taxonomic grouping in Table 1.
Nucleotide–haplotype diversity relationship based on population-level estimates (n=127) derived from a homologous 456-base region of cox1. Fitted model π=0.0081h2 shown as a thick dark line within two thin lines that represent 95% confidence intervals. The 75th and 95th percentiles of the square root transformed residual data are shown as dashed and dotted lines, respectively. Population-level estimates with the largest deviations are numbered to identify the species: 1, C. nucula; 2, E. superba; 3, M. squamiger; 4, P. zostericola; 5, R. denticulata.
The relationship between the population-level estimates of h and π was positive and non-linear (Figure 3). Model selection using AIC criteria did not reveal strong statistical differences between a simple π=βh2 model, an exponential model π=αeβh and more complex square forms involving more parameters and terms (π=α+βh2; π=α+γh+βh2). BIC criteria identified π=h2 as the most parsimonious model. The fit of this was π=0.0081h2, adjusted R2=0.823, P≪0.0001. Deviation boundaries from the fitted model, calculated using the transformed residuals, were ±0.0021+0.0081h2 for the 75th percentile boundaries and ±0.0036 +0.0081h2 for the 95th percentile boundaries. The largest deviations from the fitted model were due to samples with a higher than expected π for a given h (Figure 3; see also Table 1, Figure 2). The outlier sample of R. denticulata occurred well beyond the upper 95th percentile, whereas some samples of the chicken-liver sponge Chondrilla nucula, solitary ascidian Microcosmus squamiger and E. superba exhibited less extreme deviations just beyond the 95th boundary. One sample from the species P. zostericola occurred on the 95th percentile boundary.
Sample sizes of n⩽5 individuals produced overlapping estimates of h and π between the ‘low-diversity’ B. ardens and the ‘high-diversity’ E. superba population-level data sets (Figure 4). Samples of n=23 individuals from B. ardens and n=15 individuals from E. superba were required before the variance between subsamples was reduced to <0.25 of the total subsampling variation found in these species. For h, variance was larger among subsamples of B. ardens than among subsamples of E. superba. In contrast, for π the variance among subsamples was larger for E. superba than for B. ardens.
Sample size–cox1 diversity estimate relationships for h and π under low-diversity (Bombus ardens; black cross series) and high-diversity (Euphausia superba; gray plus series) scenarios. Each series of estimates from n=2 to n=49 comprises 100 random subsamples taken from the total sample size of 50. High–low bars indicate s.d. output from DNASP for n=50.
Discussion
Data archiving and data set re-assembly
More than half of the population-level cox1 sequence data sets from peer-reviewed publications matching our selection criteria could not be re-assembled (Figure 1). Such a high proportion of unavailable raw data impedes what can be achieved through re-analysis, which is particularly important for commonly sequenced genes like cox1, and highlights the importance of current efforts by the research community (data generators, data managers and publishers) to promote data archiving (Whitlock et al., 2010). Of those data sets that were re-assembled, 15% generated results that differed from results presented in the corresponding publications (Figure 1). This level of incompatibility is troubling. It implies that the research community also needs to promote measures to verify the credibility of published diversity estimates, archived data and references to this data.
The reduction in total sample size from 361 to 127 during data acquisition (Figure 1) begs the question of whether it is worthwhile re-assembling DNA sequence data sets for comparative analysis. To investigate whether this procedure improved our analysis, we re-modeled the relationship between h and π using our initial estimates of these metrics that were derived from the original variable-length cox1 sequences (n=127). Although the model and fit was similar (π=0.0074h2, adjusted R2=0.818, P≪0.0001), the position of notable samples from the species C. nucula and M. squamiger relative to the deviation percentile boundaries changed. These outliers occurred above the upper 95th percentile in analysis using the homologous 456-base sequences (Figure 3), but they occurred below the re-derived upper 95th percentile when using the original variable-length cox1 sequences (data not shown). This suggests that data set re-assembly and the derivation of new diversity estimates using a homologous region of DNA had increased our ability to detect subtle outliers. We recommend this approach for the comparative analysis of population-level cox1 h and π to remove the impact of variable sequence length-based error. This method also has the benefit of reducing error that may arise through the direct analysis of inaccurately published diversity estimates.
A pitfall of the manual data set re-assembly adopted herein is that it is laborious. This process could potentially be automated by adapting the methods used in meta-analyses by Bazin et al. (2006) or Wares (2010) in which population-level cox1 sequences were collated into species-level data sets. Algorithms would need to be developed to account for archived lists of unique haplotypes and to group DNA sequences according to population identifiers. It is crucial to deal with unique haplotype lists because a failure to do so would generate flawed data sets that yield biased estimates of h and π. For this type of automated approach to work, it may also be necessary to apply complimentary changes to data archives and future archiving procedures.
Haplotype and nucleotide diversity variation
Intra-specific variation for estimates of h and π was generally large compared with the variance of the total data set (Table 1, Figure 2). This finding reflects the dominant role of ecological and population-level processes in determining levels of genetic variation (for example, P. zostericola; Haig et al., 2010) in addition to underlying factors such as mutation rate (Boyer et al., 2007). However, a substantial component of the variance may also be due to error from under-sampling (see below), the inadvertent inclusion of cryptic species or possibly contaminating numt DNA/sequencing errors. The large variance among population-level samples described here implies that, even when intra-specific variance is low (for example, E. superba; Figure 2), it is important to sample species distributions thoroughly and carefully before drawing conclusions about the level of h and π for species as a whole.
In order to improve the clarity and consistency with which population-level estimates of h and π are reported in the future, we advocate comparing them against the values in Table 1. In addition to quantitative comparisons against specific values, median values derived from the complete data set can be used as a cutoff for qualitative descriptions of low and high diversity. For example, in the study by Raupach et al. (2010) on benthic shrimp species, estimates of h and π for samples of Chorismus antarcticus fall below the median value and thus could be categorized as low diversity, whereas samples of Nematocarcinus lanceopes fall above the median and thus could be termed high diversity (Table 1, Figure 2).
The relationship between h and π
The positive relationship we described between population-level estimates of cox1 h and π (Figure 3) has been noted previously by Bird et al. (2007). However, to our knowledge, this is the first time that a model describing the relationship between the two metrics has been reported. The BIC criteria identified π=h2 as the most parsimonious model. This relationship emerges because π is based on the pairwise comparison of nucleotide differences within distinct haplotypes and is dominated by the square term (Nei and Li, 1979; Nei, 1987). We propose that the deviation of individual samples from this relationship can be used to identify outliers that might not be evident through the assessment of h or π alone. The extent of deviation from the model can be assessed by comparison with the 75th and 95th percentile deviation boundaries (Figure 3). To assess the deviation of new diversity estimates, h and π should be derived from the 456-base region of cox1 defined in the Materials and methods. A predicted nucleotide diversity (πp) can be calculated from the observed haplotype diversity (ho) using the relationship πp=0.0081ho2. A value for comparison against the percentile curves (75 and 95%) for the transformed residuals can then be calculated using Δ=√∣(πo−πp)∣. The greater the deviation of a sample from the model's expectations, the greater the need for additional investigation, as this may reveal notable biological processes or the impact of methodological error.
The extreme deviation of the R. denticulata sample well beyond the upper 95th percentile in Figure 3 could reflect a range of processes according to Boyer et al. (2007), including secondary contact between divergent haplotypes, high mutation rates and the inadvertent sampling of cryptic species. The more subtle deviations found in samples of C. nucula and M. squamiger may represent secondary contact between divergent lineages (Duran and Rützler, 2006; Rius et al., 2008), whereas that shown by E. superba probably reflects the exceptional abundance and mixing of swarms of this species (Goodall-Copestake et al., 2010). An explanation for the slight deviation of the P. zostericola sample is not obvious on the basis of the corresponding publication (Haig et al., 2010). It may represent a biological process or possibly sampling error given the number of individuals involved (n=15) and the potential for error associated with a sample of this size (see Figure 4).
The effect of sample size on haplotype and nucleotide diversities
Subsampling analysis indicated that samples of n⩽5 individuals, which are not uncommon in published studies, could generate highly variable estimates that hinder accurate h and π comparisons (Figure 4). Higher sample sizes (n>5) were shown to be sufficient to discriminate low-diversity populations such as in B. ardens (Kim et al., 2009) from high-diversity populations like those in E. superba (Goodall-Copestake et al., 2010). The impact of under-sampling on estimates of h and π was context-dependent. Estimates of h were more sensitive to under-sampling bias if the focal population harbored limited cox1 variation (B. ardens), because a large number of individuals must be sampled from the population in order to register the presence of low-frequency alleles. By contrast, π was more biased by low sample numbers if the focal population was highly variable at cox1 (E. superba) as divergent alleles in such populations have a large impact on estimates derived from small sample sizes.
Results from the B. ardens and E. superba analyses showed that, with a sample size of n⩾23 individuals, the total subsampling variance was reduced by at least a factor of four. At this arbitrary level, multiple ‘broad’ categories of h and π can be distinguished; although clearly much larger sample sizes are required to achieve a greater resolution (for example, to recover non-zero values of h in B. ardens). As a rule-of-thumb we suggest using sample sizes of n⩾25 individuals in future comparisons of population-level cox1 diversity. This number is achievable for many species and represents a practical compromise between the current expense of data generation and the obvious benefit of high n numbers.
Significance beyond cox1
The present study highlights a number of issues concerning the archiving and comparative analysis of population-level cox1 sequence data, on the basis of which we make a number of recommendations. In addition, we propose a general method to assess paired haplotype–nucleotide diversity estimates. This is not only relevant to cox1 data but also applicable to other sources of haploid (chloroplast, mitochondrial, sex-chromosome) DNA sequence data used to investigate intra-specific diversity in both animals and plants. In particular, we recommend that a similar approach is taken for analyses of mitochondrial control region (D-loop) data, which is widely used for population-level studies on mammals.
Data archiving
There were no data to deposit.
Accession codes
References
Arruda CCB, Beasley CR, Vallinoto M, Marques-Silva NS, Tagliaro CH (2009). Significant genetic differentiation among populations of Anomalocardia brasiliana (Gmelin, 1791): A bivalve with planktonic larval dispersion. Genet Molec Biol 32: 423–430.
Bazin E, Glémin S, Galtier N (2006). Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572.
Bucklin A, Steinke D, Blanco-Bercial L (2011). DNA Barcoding of Marine Metazoa. Annu Rev Mar Sci 3: 471–508.
Burnham KP, Anderson DR (2002). Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer: New York.
Bird CE, Holland BS, Bowen BW, Toonen RJ (2007). Contrasting phylogeography in three endemic Hawaiian limpets (Cellana spp.) with similar life histories. Mol Ecol 16: 3173–3186.
Boyer SL, Baker JM, Giribet G (2007). Deep genetic divergences in Aoraki denticulata (arachnida, opiliones, cyphophthalmi): a widespread ‘mite harvestman’ defies DNA taxonomy. Mol Ecol 16: 4999–5016.
Cárdenas L, Castilla JC, Viard F (2009). A phylogeographical analysis across three biogeographical provinces of the south-eastern Pacific: the case of the marine gastropod Concholepas concholepas. J Biogeogr 36: 969–981.
Clark AG, Whittam TS (1992). Sequencing errors and molecular evolutionary analysis. Mol Biol Evol 9: 744–752.
Duran S, Rützler K (2006). Ecological speciation in a Caribbean marine sponge. Mol Phylogent Evol 40: 292–297.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–297.
Fu YX (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925.
Goodall-Copestake WP, Pérez-Espona S, Clark MS, Murphy EJ, Seear PJ, Tarling GA (2010). Swarms of diversity at the gene cox1 in Antarctic krill. Heredity 104: 513–518.
Haig JA, Connolly RM, Hughes JM (2010). Little shrimp left on the shelf: the roles that sea-level change, ocean currents and continental shelf width play in the genetic connectivity of a seagrass-associated species. J Biogeogr 37: 1570–1583.
Jeong HC, Kim JA, Im HH, Jeong HU, Hong MY, Lee JE et al. (2009). Mitochondrial DNA Sequence Variation of the Swallowtail Butterfly, Papilio xuthus, and the Cabbage Butterfly, Pieris rapae. Biochem Genet 47: 165–178.
Kim MJ, Yoon HJ, Im HH, Jeong HU, Kim MI, Kim SR et al. (2009). Mitochondrial DNA sequence variation of the bumblebee, Bombus ardens (Hymenoptera: Apidae). J Asia Pacific Entomol 12: 133–139.
Knowlton N (1986). Cryptic and sibling species among the decapod Crustacea. J Crustac Biol 6: 356–363.
Kojima S, Ozeki S, Iijima A, Okoshi K, Suzuki T, Hayashi I et al. (2008). Genetic characteristics of three recently discovered populations of the tideland snail Cerithidea djadjariensis (Martin) (Mollusca, Gastropoda) from the Pacific coast of the eastern Japan. Plankton Benthos Res 3: 96–100.
Lara-Ruiz P, Chiarello AG, Santos FR (2008). Extreme population divergence and conservation implications for the rare endangered Atlantic Forest sloth, Bradypus torquatus (Pilosa: Bradypodidae). Biol Cons 141: 1332–1342.
Lejeusne C, Chevaldonné P (2006). Brooding crustaceans in a highly fragmented habitat: the genetic structure of Mediterranean marine cave-dwelling mysid populations. Mol Ecol 15: 4123–4140.
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ (1994). Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39: 174–190.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Nei M, Li WH (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sct USA 76: 5269–5273.
Palero F, Abelló P, Macpherson E, Gristina M, Pascual M (2008). Phylogeography of the European spiny lobster (Palinurus elephas): influence of current oceanographical features and historical processes. Mol Phylogenet Evol 48: 708–717.
Pellerito R, Arculeo M, Bonhomme F (2009). Recent expansion of Northeast Atlantic and Mediterranean populations of Melicertus (Penaeus) kerathurus (Crustacea: Decapoda). Fish Sci 75: 1089–1095.
Raupach MJ, Thatje S, Dambach J, Rehm P, Misof B, Leese F (2010). Genetic homogeneity and circum-Antarctic distribution of two benthic shrimp species of the Southern Ocean, Chorismus antarcticus and Nematocarcinus lanceopes. Mar Biol 157: 1783–1797.
Ribeiro PA, Branco M, Hawkins SJ, Santos AM (2010). Recent changes in the distribution of a marine gastropod, Patella rustica, across the Iberian Atlantic coast did not result in diminished genetic diversity or increased connectivity. J Biogeogr 37: 1782–1796.
Rius M, Pascual M, Turon X (2008). Phylogeography of the widespread marine invader Microcosmus squamiger (Ascidiacea) reveals high genetic diversity of introduced populations and non-independent colonizations. Diversity Distrib 14: 818–828.
Santos SR (2006). Patterns of genetic connectivity among anchialine habitats: a case study of the endemic Hawaiian shrimp Halocaridina rubra on the island of Hawaii. Mol Ecol 15: 2699–2718.
Strasser CA, Barber PH (2009). Limited genetic variation and structure in softshell clams (Mya arenaria) across their native and introduced range. Conserv Genet 10: 803–814.
Tajima F (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Wares JP (2010). Natural distributions of mitochondrial sequence diversity support new null hypotheses. Evolution 64: 1136–1142.
Whitlock MC, McPeek MA, Rausher MD, Rieseberg L, Moore AJ (2010). Data archiving. Am Nat 175: 145–146.
Acknowledgements
We thank the scientists responsible for generating the DNA sequence accessions used in this study, authors responding to queries about their data, Sílvia Pérez-Espona for fruitful discussions and three anonymous referees for their comments on the manuscript. We carried out this work as part of the Ecosystems group within British Antarctic Survey Polar Science for Planet Earth Programme funded by the Natural Environment Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Goodall-Copestake, W., Tarling, G. & Murphy, E. On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity 109, 50–56 (2012). https://doi.org/10.1038/hdy.2012.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.12
Keywords
This article is cited by
-
A comprehensive DNA barcoding of Indian freshwater fishes of the Indus River system, Beas
Scientific Reports (2024)
-
Genetic diversity and population differentiation in Earliella scabrosa, a pantropical species of Polyporales
Scientific Reports (2023)
-
Whole mitochondrial genomes reveal the relatedness of the browsing ant incursions in Australia
Scientific Reports (2023)
-
Mitochondrial DNA diversity of the eastern black rhinoceros (Diceros bicornis michaeli) in Tanzania: implications for future conservation
Conservation Genetics (2023)
-
Comparative phylogeography and demographic histories of five widely distributed tropical eastern Pacific fishes
Marine Biology (2023)