Abstract
Continental islands offer an excellent opportunity to investigate adaptive processes and to time microevolutionary changes that precede macroevolutionary events. We performed a population genetic study of the fire salamander (Salamandra salamandra), a species that displays unique intraspecific diversity of reproductive strategies, to address the microevolutionary processes leading to phenotypic and genetic differentiation of island, coastal and interior populations. We used eight microsatellite markers to estimate genetic diversity, population structure and demographic parameters in viviparous insular populations and ovoviviparous coastal and interior populations. Our results show considerable genetic differentiation (FST range: 0.06–0.27), and no clear signs of gene flow among populations, except between the large and admixed interior populations. We find no support for island colonization by rafting or intentional/accidental anthropogenic introductions, indicating that rising sea levels were responsible for isolation of the island populations approximately 9000 years ago. Our study provides evidence of rapid genetic differentiation between island and coastal populations, and rapid evolution of viviparity driven by climatic selective pressures on island populations, geographic isolation with genetic drift, or a combination of these factors. Studies of these viviparous island populations in early stages of divergence help us better understand the microevolutionary processes involved in rapid phenotypic shifts.
Similar content being viewed by others
Introduction
Adaptation and diversification of insular populations have been an important source of evidence for evolution (Grant, 1998; Losos and Ricklefs, 2009). Islands are natural laboratories for evolutionary studies because their intrinsic characteristics such as relative age, geographic isolation, smaller population sizes and reduced number of species, help us isolate potential mechanisms underlying species formation and adaptation to local environments. In young islands, such as continental islands that become isolated from the mainland due to relatively recent increases in sea level, evolutionary processes are occurring over contemporary timescales allowing us to infer the phenotypic and genetic consequences of isolation and incipient divergence (Thompson, 1998; Stockwell et al., 2003).
Examples of evolutionary change in the fauna and flora endemic to oceanic islands are abundant because colonization occurred a long time ago and adaptive radiations have often occurred into new ecological niches (Emerson, 2002). Larger and more isolated islands tend to harbor more endemic species than small islands because they offer more habitats (MacArthur and Wilson, 1967), provide ecological gradients and habitat diversity (Lovette et al., 2002) and support larger population sizes (Gavrilets and Vose, 2005), all of which promote speciation and prevent loss of newly evolved forms due to stochastic extinction. In contrast, evidence for evolutionary changes in more recent continental islands isolated during the Holocene (7000–10 000 years ago) is far less common. Species endemic to continental islands arise via isolation rather than independent colonization from the mainland, and most continental islands are only recently isolated (in some cases only thousands of years). In addition, continental islands tend to be smaller in size, close to the coastline, and have ecological niches similar to those on the adjacent mainland, which leads to lower overall diversification, but offers important insights into recent evolutionary processes.
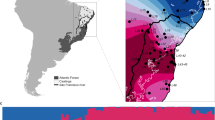
The European fire salamander (Salamandra salamandra) ranges from the southwestern Iberian Peninsula to Ukraine and Greece at the easternmost extent of its range (Montori and Herrero, 2004). Populations of fire salamanders are polymorphic in reproductive mode; most populations are ovoviviparous, with females giving birth to larvae that develop in aquatic habitats. However, a few populations throughout the range are viviparous, with females giving birth to terrestrial, fully metamorphosed individuals (Dopazo and Alberch, 1994; Alcobendas et al., 1996; Buckley et al., 2007). Viviparous populations occur only in the Cantabrian mountains and Cantabrian coast (central-northern Iberia; S. s. bernardezi), the southwest Pyrenees (S. s. fastuosa) and on two small offshore islands (San Martiño and Ons) in the Atlantic ocean, 3.6 and 6 km from the northwestern coast of Spain, respectively (S. s. gallaica; Velo-Antón et al., 2007; Figure 1). Transitions from ovoviviparity to viviparity have occurred repeatedly in the three amphibian orders (Wake, 2004), and viviparity is a derived reproductive mode in salamanders (Veith et al., 1998; García-París et al., 2003; Weisrock et al., 2006). An early phylogenetic and phylogeographic study using mitochondrial and nuclear DNA markers indicated that viviparity arose once within S. salamandra in the Cantabrian Mountains (northern Iberian Peninsula; S. s. bernardezi), and spread along the coast to the Pyrenees with the evolution of S. s. fastuosa (García-París et al., 2003). However, a more recent phylogenetic study including the Atlantic island viviparous populations showed that S. s. gallaica have independently evolved viviparity (Velo-Antón et al., 2007), and these island forms also differ from the other two viviparous subspecies in coloration pattern, body shape and size.
Sampling design and distribution of Iberian S. salamandra subspecies. The ranges of Iberian S. salamandra subspecies are represented on the map on the right. S. s. bernardezi and western populations of S. s. fastuosa are viviparous. The map on the left shows sampling localities included in this study. Ovoviviparous populations are represented by black circles and viviparous populations by black stars.
In this study we examine the microevolutionary processes leading to reproductive and genetic differentiation of coastal and island populations of S. salamandra by surveying genetic diversity, population structure and demographic parameters in viviparous insular populations and ovoviviparous coastal and interior populations. We propose two hypotheses to explain the origin of insular populations: (1) Present insular populations were isolated when coastal mountaintops became islands due to rising sea levels approximately 9000 years ago. Under this scenario we expect to find high genetic differentiation with low or no signals of gene flow between island and mainland (coastal and interior) populations; (2) Island viviparous populations were established by seawater colonization or intentional/accidental anthropogenic introductions from mainland populations. In this case, we expect to find low genetic divergence and genetic admixture between island and mainland populations. We also evaluate the effects of founder population sizes (either by geographic isolation or oversea colonization) on demographics, genetic diversity and differentiation in both islands and discuss potential factors for the evolution of viviparity in island populations.
We analyzed genetic diversity and relationships among the two viviparous island populations, three historical island ovoviviparous populations, now connected to mainland, and two interior ovoviviparous populations, and used the distribution and degree of genetic differentiation to infer the microevolutionary processes acting on populations. Population genetic studies of island species in early stages of divergence reveal evolutionary processes affecting phenotypic and genetic changes at short timescales. Thus, understanding the evolutionary history of the viviparous island populations of S. salamandra will also provide an ideal framework for the study of the ecological and evolutionary processes involved in rapid shifts in reproductive mode.
Methods
Study sites and population sampling
Our study includes samples from the two insular viviparous populations of S. salamandra (San Martiño and Ons), three ovoviviparous geographically adjacent coastal populations (Monteferro, Melide and Grove) and two ovoviviparous interior populations (Saramagal and Lourizán; Figure 1). These ovoviviparous coastal and interior populations are likely the closest relatives to island viviparous populations based on morphological similarities (S. s. gallaica phenotype), previous mitochondrial DNA results (Velo-Antón et al., 2007) and geographic proximity. San Martiño and Ons islands are part of the Galician Atlantic Islands National Park, and are separated by 3.6 and 6 km from the mainland, respectively, and 12 km from each other. Both islands are relatively small: San Martiño is 2.3 km long and 1 km wide (146 ha), and Ons is 5 km long and 1.2 km wide (428 ha). The three coastal populations (Grove, Melide and Monteferro) are located on small peninsulas, and each is connected to the mainland by a low elevation isthmus (1, 25 and 30 m a.s.l., respectively). A continuous and active sedimentation processes began 2000 years ago along this shore (Dias et al., 2000) and might have reconnected these historical island populations to the continent (Figure 2). These are currently the geographically closest continental populations (5–10 km) from the islands. The interior populations (Saramagal and Lourizán) are separated by 10–25 km from their closest coastal populations (Figure 1).
Population structure and gene flow in focal populations of S. salamandra. The six genetic demes inferred in STRUCTURE are represented with different colors. Individual assignment to the six genetic demes is shown for each population (bottom panel). Blue arrows on the map identify the location of peninsular isthmuses and their elevation, likely resulting from sedimentation processes along the shore in the last 2000–3000 years.
We searched for salamanders during diurnal and nocturnal samplings in each population. We collected a mean of 32 tissue samples per locality (range 28–36) from adult salamanders (tail or toe clips), and preserved them in 100% ethanol for later DNA analyses (Table 1).
DNA extraction and genotyping
Tissues were digested in (200 μl) lysis buffer with 1 μl Proteinase K (25 mg ml−1), and genomic DNA was purified using a lithium chloride protocol (Gemmell and Akiyama, 1996). The resulting DNA pellets were air dried and resuspended in 50 μl TE, pH 8. Genomic DNA was diluted to a final concentration of 50 ng μl−1 and used as template in PCR amplifications of eight microsatellite loci (E2, E6, E7, E8, E11, E14, S3 and S23; Steinfartz et al., 2004). Forward primers were labeled with 5′-fluorescent tags (6-FAM, HEX or NED) for visualization. PCR amplifications were performed in reactions of 12 μl total volume including 1 μl of DNA template (∼100 ng μl−1), 1.5 μl 10 × PCR buffer (100 mM Tris-HCl, 15 mM MgCl2, 500 mM KCl) MgCl2, 0.5 μl dNTPs (25 mM), 0.5 μl forward and reverse primers (10 mM) and 0.05 μl Taq polymerase (5 U μl−1). We amplified loci individually and ran PCR products in three multiplex reactions on an ABI 3100 (Applied Biosystems, Foster, CA, USA). Fragments were sized with ROX-500. Results were scored and binned using GeneMapper version 3.7 (Applied Biosystems).
Genetic data analyses
We used MICRO-CHECKER 2.2.3 to detect potential null alleles and scoring errors due to stuttering or large allele dropout (Van Oosterhout et al., 2004). We tested for significant deviations from Hardy–Weinberg equilibrium and the presence of linkage disequilibrium using ARLEQUIN 3.11 (Excoffier et al., 2005) and tested for significance using a Markov chain method with 10 000 dememorization steps and 1000 batches of 10 000 iterations per batch and sequential Bonferroni correction (Rice, 1989) for multiple comparisons.
We used the software GENALEX v.6 (Peakall and Smouse, 2006) to calculate indices of population genetic diversity (observed heterozygosity, HO; expected heterozygosity, HE; mean number of alleles, NA and allelic richness AR), and estimate genetic relatedness, r, within populations (Queller and Goodnight, 1989). Mean pairwise relatedness values were calculated and compared with the confidence intervals (CIs) obtained by bootstrapping over genotypes within populations. The program HP-RARE (Kalinowski, 2005) was used to compute estimates of unbiased allelic richness and private allelic richness.
Genetic differentiation between populations was calculated using pairwise FST values (Weir and Cockerham, 1984) in ARLEQUIN 3.11, and significance of pairwise comparisons tested using an exact test (Goudet et al., 1996) with 1000 iterations, and P-values adjusted using the sequential Bonferroni correction (Rice, 1989). We also used pairwise ϕ′ST (999 bootstraps) (Meirmans, 2006) to account for potential differences in effective population sizes among our populations using the software GENODIVE v2.09b (Meirmans and Van Tienderen, 2004). ϕ′ST is an AMOVA analog of FST and is standardized by dividing the observed ϕ′ST by the theoretical maximum given the present within population variance in allele frequencies.
To detect potential loss of genetic diversity due to founder events, we tested for differences between each island and their genetically closest coastal populations (based on FST and ϕ′ST values, Table 2). We used non-parametric Mann–Whitney U-tests to compare the mean number of alleles (NA) and expected heterozygosity (HE) for each locus and pair of populations.
We used Bayesian assignment tests to infer the number of genetic demes present in our sample. The program STRUCTURE 2.3.1 (Pritchard et al., 2000) was run 20 times on individual multilocus microsatellite genotypes for a number of genetic clusters or demes (K) ranging from 1 to 7. We implemented an admixture model with uncorrelated allele frequencies and without prior information on sample population membership, a burn-in period of 500 000, and a run length of 106 iterations. The most likely value for K was estimated by the highest value of log probability of data L(K) (Falush et al., 2003), and the Δ(K), a measure based on the second order rate of change in L(K) (Evanno et al., 2005). We used the program CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007) to align cluster membership coefficients from the 20 replicate cluster analyses from each K-value chosen using the LargeKGreedy algorithm.
We used LAMARC 2.1.3 (Kuhner, 2006) to estimate effective population sizes (Θ) and growth rates (G) of our seven focal populations. Owing to computational constraints, we used a Brownian-motion microsatellite model (an approximation of the stepwise model), for all loci to decrease the time of the runs. We used the Bayesian method with 500 000 final recorded parameter sets with a burn-in of 100 000 steps, sampling every 100 steps, and five replicates for each analysis.
We estimated migration rates, M (M=m/μ, where m is the immigration rate per generation), between populations using a coalescent based Monte Carlo Markov Chain method implemented in MIGRATE-N 3.2.7 (Beerli and Felsenstein, 1999). We analyzed the microsatellite data set under a Brownian approximation model and mutation was considered to be constant for all loci. We used FST estimates and a UPGMA tree as starting parameters for the estimation of M and performed five independent runs using one long chain with a run of 1 × 107 recorded parameter genealogies after discarding the first 1 × 105 genealogies as burn-in for each locus. Both LAMARC and MIGRATE estimated approximate CIs around its maximum probable estimate for each parameter. Parameter convergence was verified by examining the stationarity in parameter trends over the length of the chains and the effective sample sizes obtained for each Θ, G and migration rate using TRACER 1.5 (http://evolve.zoo.ox.ac.uk).
To test for evidence of historical genetic bottlenecks we used the M-ratio test (Garza and Williamson, 2001), which uses the M statistic (the ratio of the number of alleles to the range of allele sizes) to detect reductions in effective population size for a sample of microsatellite loci. This method considers that quantitative diversity (frequency of alleles and total number of alleles, k) is reduced more quickly than spatial diversity (distance between alleles and overall range in allele size: r) in bottlenecked populations. We generated a critical value (Mc) at α=0.05 to test estimates of M for evidence of a bottleneck using 10 000 permutations and conservative parameters for two-phased model (percentage of mutations greater than one step, Δg=2.8; average size of a non-one-step mutation, r=0.12; and different effective population sizes, θ=1, 2 and 10). We also used the software BOTTLENECK (Piry et al., 1999) to test for recent population bottleneck events. This program tests for reductions in effective population size by considering that alleles are generally lost faster than heterozygosity (Hedrick and Cockerham, 1986) and thus, populations that have experienced a reduction in Ne are expected to have excess heterozygosity (HE) relative to that expected under mutation-drift equilibrium (Heq) (Cornuet and Luikart, 1996). Following the recommendations of the program for limited sample size, we used the Wilcoxon signed-rank test under two mutation models: the stepwise mutation model and the two-phased model with 70% stepwise mutation model, using 100 000 replicates. We also examined the distribution of allele frequencies among all the populations to determine whether a mode shift characteristic of a bottleneck had occurred (Luikart and Cornuet, 1998).
Results
All microsatellites were highly polymorphic across the seven populations analyzed, and number of alleles ranged from 10 (loci E8 and E11) to 38 (E2) with a mean of 16 alleles per locus. Sample sizes genotyped across our seven focal populations ranged from 28 to 36 (Table 1). No evidence of null alleles, large allele dropouts or stuttering was detected at the 99% confidence level across all loci. After sequential Bonferroni corrections, comparisons among all pairs of loci over all populations revealed no linkage disequilibrium, and significant deviations from Hardy–Weinberg equilibrium at the 99% confidence interval were detected for a single locus (E8), and only in the Ons and Grove populations.
Highest genetic diversities (HO, HE, NA and AR) were found in interior populations (Lourizán and Saramagal); coastal populations and the large island population (Ons) showed intermediate values, and the smallest island population (San Martiño) showed the lowest genetic diversity indexes (Table 1). Relatedness values (r) corroborated these results with the highest level of inbreeding for San Martiño population (r=0.47, close to full sibs) and the lowest for interior populations (Lourizán, r=0.04; and Saramagal, r=0.01; Table 1). The population with highest private allelic richness was the island population of Ons, followed by the interior populations. San Martiño population showed the lowest private allelic richness (Table 1). We found significant differences in genetic diversity values between San Martiño and Melide populations (NA, z=−2.26, P=0.01; and HE, z=−1.73, P=0.04) but not between Ons and Melide (NA, z=−0.11, P=0.45; and HE, z=−0.05, P=0.49).
All pairwise population FST comparisons were significant and ranged from 0.014 (between interior populations) to 0.287 (between San Martiño and the coastal population Monteferro) (Table 2). Island populations showed lower genetic divergences when compared with interior populations (FST=0.107–0.178) than coastal populations (FST=0.152–0.287). All pairwise ϕ′ST comparisons were also significant and showed similar genetic patterns, with interior populations less differentiated from each other (ϕ′ST=0.017) and high genetic differentiation between island and mainland populations (ϕ′ST=0.168–0.298).
Assignment tests separated the sampled populations into six genetic clusters (K=6), based on the log probability of data L(K), and the Evanno criterion for number of demes, Δ(K). Both methods separated island and coastal populations into independent clusters and grouped the two interior populations (Figure 2). Assignment tests implemented by STRUCTURE showed the proportion of membership (Q) for each defined deme was higher than 93% in island and coastal populations, and 75% and 81% for interior populations (Lourizán and Saramagal, respectively).
LAMARC analyses recovered similar effective population size (Θ) in island and coastal populations, but significantly higher values for both interior populations (Figure 3). We translated Θ estimates to average effective population size for each study population (assuming a microsatellite mutation rate of 10−3 per locus per generation reported for Ambystoma tigrinum; Bulut et al., 2009; Table 1). Maximum probable estimate values of population growth were slightly different from zero, either with slightly negative growth rate in island and coastal populations, or positive growth rate in interior populations, but CIs for all those estimates (CIs) overlapped across populations (Figure 3).
Immigration rates estimated from MIGRATE showed significant and symmetrical gene flow only between interior populations (Lourizán and Saramagal). However, lower immigration-rate 95% CIs between island and coastal populations did not depart from zero, suggesting a likely absence of gene flow between those populations.
We found evidence of historical bottlenecks in island and coastal populations using M-ratio tests. The 95% threshold value, Mc, ranged from 0.72 to 0.77 under different effective population sizes (θ=1, 2 and 10), and all island and coastal populations showed lower mean M values for each θ (Table 4). None of the interior populations showed significant evidence of reduction in Ne and mean M values were higher than Mc values for each θ. For estimates of recent bottlenecks (excess of heterozygosity), we only found significant values under the two-phased model (P<0.05) for interior populations (Lourizán and Saramagal) but not for island or coastal populations (Table 4).
Discussion
Island isolation vs seawater colonization
Our study has implications for the origin of insular populations of S. salamandra along the northwestern Iberian coast. In general, the presence of amphibians on islands can be explained by (1) population isolation before island formation and (2) overseas colonization or intentional/accidental anthropogenic introductions from adjacent mainland populations. Amphibian tolerance to seawater varies among species, and long-distance movements by rafting have been proposed as a possible mechanisms of colonization for continental islands (Hedges et al., 1992; Vences et al., 2003; Measey et al., 2007), suggesting that seawater is not necessarily an absolute barrier to gene flow. In this study we found high levels of genetic differentiation and absence of gene flow between island and coastal populations (FST=0.181–0.287, Tables 2 and 3; Figure 2). We found no evidence of colonization by rafting from the coast or intentional/accidental anthropogenic introductions. Therefore, our study supports the hypothesis that island populations of S. salamandra (San Martiño and Ons, Figure 1) derived from ancestral populations that occupied the coastal highlands before they became isolated from the mainland with rising sea levels approximately 8000–9000 years ago (Dias et al., 2000). The fact that isolated populations do not seem to have experienced any subsequent migration, indicates that sea water is an effective barrier to dispersal and gene flow for S. salamandra, even over short overseas distances (3.6–6 km). The same pattern was recently detected for populations of slender salamanders, Batrachoseps, on the islands of San Francisco Bay (Martínez-Solano and Lawson, 2009), which share similar age (9000 years) and distances among islands and mainland.
A third possible scenario to explain the genetic divergence found between these populations is one of isolation and incipient genetic differentiation by genetic drift before the formation of the islands. Although, with the data at hand, population isolation pre- or post-island formation cannot be distinguished, our data point out to genetic differentiation facilitated by restriction of gene flow between populations after island formation, as only distant interior populations are not differentiated (Figure 2).
Population genetics of island and mainland S. s. gallaica
Our study shows remarkable genetic differentiation among sampled populations given the small geographic scale of this study (Table 2; Figure 2). The FST values obtained were higher than those documented in other fine-scale salamander studies (Cabe et al., 2007; Zamudio and Wieczorek, 2007; Purrenhage et al., 2009) and frog island populations (Lind et al., 2010), but showed similar levels of divergence to other amphibian population genetic studies covering larger regions (Spear et al., 2005; Savage et al., 2010). The similarities in genetic diversity and demographics among island and coastal populations (Table 1; Figure 3) underscore their similar histories, despite the current differences in connectivity to the mainland.
The shape of the coastline of western Iberia has changed significantly since the Last Glacial Maximum due to sea level rising until the mid Holocene (between 5 to 2.5 Ky BP) and continental erosion and sedimentation processes in estuaries over the last 2000–3000 years (Dias et al., 2000). The erosion process occurred due to natural factors and historical anthropogenic activities, such as deforestation and land cultivation, that contributed to the positive sedimentary balance along the western Iberian shore (Dias et al., 2000), connecting islands to mainland (Calado, 1994) and forming sand-barriers along the coast (for example, Cíes barrier-lagoon; Costas et al., 2009). Therefore, the three focal coastal populations located on small coastal peninsulas were likely continental islands that reconnected to the continent by sedimentation processes in the last thousand years (Dias et al., 2000; Figure 2). Historical geographic isolation on these islands would explain the unexpected high genetic differentiation, absence of gene flow, and the similarities of genetic diversity and demography with island populations. Island and coastal populations have less genetic diversity and lower historical population sizes and growth rates than interior populations (Table 1 and Figure 3), and they also differ in historical and recent demographic patterns (Table 4).
The fact that island and coastal populations are similarly differentiated compared with island and interior populations (Table 2) might also be explained by a longer mutation—genetic drift scenario in island populations, because the isolation by sea level changes occurred later for the coastal populations. The following results support isolation and high genetic drift as mechanisms for the observed divergence between island and coastal populations: (1) island and coastal populations are more closely related than island and interior populations at mitochondrial DNA markers (Velo-Antón et al., 2007); and (2) divergences between islands are higher than between island and interior populations (Table 2). Moreover, extreme genetic drift in island populations might be facilitated by the small effective population size of founder populations on small islands.
Effect of founder population sizes on islands
Founder events are important mechanisms promoting evolutionary change on islands (Mayr, 1954; Templeton, 1980; Grant, 1998). The size of a founder population is crucial for the long-term survival of island populations; small founder populations can potentially compromise survival due to loss of genetic diversity produced by drift and genetic erosion due to inbreeding (Baker and Moeed, 1987; Hartl and Clark, 1997). The founding populations of San Martiño and Ons islands may have been small because of the limited size of both islands (146 and 428 ha, respectively) and thus, it is likely that the geographic isolation of individuals from the mainland population could function as bottleneck events within each island. Common genetic signatures of population bottlenecks are the reduction of heterozygosity and allelic richness (Nei et al., 1975) and the increase of genetic differentiation from source population (Chakraborty and Nei, 1977). We found some differences in population genetic patterns between the two island populations, indicating different colonization histories and/or demographic processes since their isolation. The San Martiño population shows remarkably low genetic diversity and high values of relatedness compared with other studied populations (Table 1). In contrast, the Ons population maintains similar values of genetic diversity and relatedness as coastal populations (Table 1), even though both island and coastal populations have low genetic diversity values. Thus, our study shows a significant reduction of genetic variation in San Martiño population, likely due to a single and recent founder event that reduced population genetic variation (Mayr, 1954; Templeton, 1980; Hundertmark and Van Daele, 2010). In fact, the current small population size in San Martiño might be the result of a severe founding bottleneck in the past. In contrast, the founding of Ons population did not significantly reduce its genetic diversity. These differences may be due to the smaller size of San Martiño island (146 ha); if population densities before isolation were similar on both islands, the founding population size may have been significantly smaller than on Ons (428 ha).
Genetic isolation vs environmental factors for viviparity in island populations
Viviparity in S. salamandra is characterized by an acceleration of embryonic development (Dopazo and Alberch, 1994; Dopazo and Korenblum, 2000), the early hatching of embryos within the maternal oviducts coupled with intrauterine cannibalism, and longer retention of hatchlings in the oviducts. The acceleration of embryonic development occurs during the non-feeding lecithotrophic stages of development (S. s. bernardezi; Buckley et al., 2007), and is also facilitated by cannibalistic feeding in the post-hatching stage via oophagy, ingestion of eggs, and adelphophagy, and ingestion of larvae; (Dopazo and Alberch, 1994; Wourms, 1981; Greven, 1998). Thus, viviparity in our island populations could be determined not only through physiological and/or genetic mechanisms but also through oviductal retention of the larvae, allowing the embryos and hatchlings to grow faster and metamorphose before birth.
Genetic drift facilitates the evolutionary differentiation and adaptive processes, especially on islands (Mayr, 1954; Barton, 1998). Rapid genetic differentiation and drift could lead to the evolution of viviparity via the selection of this trait and increase of its frequency in isolated populations; however, it is unlikely that genetic drift alone led the evolution to viviparity in two island populations simultaneously. Furthermore, this reproductive mode shift could also be the result of phenotypic plasticity or adaptation to selection regimes on islands. We exclude phenotypic plasticity as a potential factor explaining viviparity on these islands because all island females exhibited viviparity when they were maintained in captivity, with water availability, until parturition (Velo-Antón et al., 2007). However, we cannot rule out a process of genetic assimilation (Waddington, 1942) on the islands. This process have been shown in colonizing island populations that initially survived due to phenotypic plasticity, which was subsequently erased by selective forces within a few thousand years (Aubret and Shine, 2009).
Adaptation to new selective regimes might also favor viviparity as a reproductive strategy in island environments. Although the strength of selection might be less effective in small populations due to low effective population sizes, studies on island populations identify directional (Clegg et al., 2002) and divergent selection (Lind et al., 2010) promoting microevolutionary changes. Our results show highly divergent island and coastal populations with similar genetic diversity and historical demographic patterns; however, viviparity occurs only in island populations, suggesting that selective pressures in island environments, rather than genetic isolation and drift alone, may have led to a rapid switch in reproductive mode.
Several environmental factors have been identified as potential selective forces for the evolution of viviparity in reptiles (Neill, 1964; Shine and Bull, 1979; Shine, 1985; Qualls and Shine, 1998). Because this evolutionary step completely removes the aquatic larval stage, the lack of water for larval development could explain this microevolutionary event in San Martiño population, but would not explain viviparity on Ons, where adequate water bodies for amphibian reproduction are common, and two additional pond-breeding amphibians occur (an anuran, Discoglossus galganoi; and a newt, Lissotriton boscai). The ‘cold climate hypothesis’ has also been proposed for the evolution of reptilian viviparity (Shine, 1985; Heulin et al., 1991; Heulin et al., 1997; Hodges, 2004). An oviparous lineage of the lizard Lerista bouganvillii occurs in south-western Australia, whereas the viviparous lineage occurs on two colder islands separated approximately 1000 km from the mainland (Qualls and Shine, 1998). However, climate in the northwestern Iberian islands is warmer and drier than on the adjacent mainland (MeteoGalicia, 2003). Furthermore, low water availability in the dry karstic limestone substrates in the Cantabrian Mountain populations of S. s. bernardezi (García-París et al., 2003) and Moroccan populations of S. algira (Beukema et al., 2010) has been identified as a potential trigger for the evolution of viviparity. Thus, we suggest that a ‘dry-climate hypothesis’ better explains the evolution to viviparity in S. salamandra. Clearly, experiments to identify the potential environmental pressures functioning as selective forces for intraspecific reproductive mode shifts in island populations of S. salamandra will be a useful next step in this system.
Our data did not allow us to distinguish whether viviparity evolved independently on each island or whether viviparous populations occurred in coastal populations before the rise of the sea level, with posterior replacement and exclusion from the continent by ovoviviparous populations. However, the ovoviviparous reproductive mode in continental populations of S. s. gallaica (Figure 1), the genetic similarities between island and coastal mitochondrial DNA haplotypes (Velo-Antón et al., 2007), and particular aspects of the insular environment suggest a rapid (within the last 9000 years) and independent change of reproductive mode for each population since their isolation.
Conservation implications
Our data show that the interior populations show higher genetic diversity and historical population sizes than island or coastal populations, yet they are facing reduction in current population sizes (Table 4), probably owing to anthropogenic habitat loss. The Lourizán population is next to a city and an industrial area, and the Saramagal population is located in a mountain area, which might be negatively affected by eucalypt plantations (Cordero Rivera et al., 2007). The absence of recent bottlenecks in island and coastal populations might be linked to the preservation of their habitats, as they are located in natural areas with restricted human and urban development. On the other hand, our results show evidence of historical reductions in population size for island and coastal populations, contributing to the reduction of genetic diversity and genetic differentiation due to drift. However, it is the viviparous island population of San Martiño that is the most threatened because of the extremely low genetic diversity and high relatedness among individuals (r=0.47; Table 1). Despite similar historical population sizes estimated in LAMARC for both island populations, the San Martiño population currently harbors only dozens of individuals, compared with hundreds or even thousands of individuals on Ons (GVA, personal observation). The San Martiño population is highly threatened and if adequate conservation measures are not implemented on this island in the near future, this could result in the loss of one of the two recently evolved viviparous populations in this species.
Conclusions
Our findings indicate that: (1) the establishment of S. salamandra populations on the islands of San Martiño and Ons occurred before the rise of sea levels, and not due to rafting or anthropogenic introductions, (2) allopatric isolation and small founder populations have led to a reduction of genetic diversity and rapid genetic differentiation by extreme genetic drift in island populations, as well as in coastal populations, which could have been continental islands as recently as 2000 years ago and (3) the rapid transition to viviparity might be driven by climatic selective pressures on island populations, geographic isolation with genetic drift or a combination of these factors.
Natural selection and genetic drift are two important forces driving microevolutionary change. Assuming that the current and historical island populations diverged by isolation and genetic drift, and knowing viviparity only occurs in the island populations, we suggest natural selection under particular island ecological pressures as the underlying factor for this reproductive shift in S. salamandra. Our study clearly highlights the utility of S. salamandra as a model for investigating rapid evolutionary change in populations with different histories and ecological settings.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad. g6d91493.
References
Alcobendas M, Dopazo H, Alberch P (1996). Geographic variation in allozymes of populations of Salamandra salamandra (Amphibia: Urodela) exhibiting distinct reproductive modes. J Evolution Biol 9: 83–102.
Aubret F, Shine R (2009). Genetic assimilation and the post-colonisation erosion of phenotypic plasticity in island Tiger snakes. Curr Biol 19: 1932–1936.
Baker AJ, Moeed A (1987). Rapid genetic differentiation and founder effect in colonizing populations of common mynas (Acridotheres tristis). Evolution 41: 525–538.
Barton NH (1998). Natural selection and random genetic drift as causes of evolution on islands. In: Grant PR (eds). Evolution on Islands. Oxford University Press: Oxford. pp 102–123.
Beerli P, Felsenstein J (1999). Maximum likelihood estimation of migration rates and effective population numbers in two populations. Genetics 152: 763–773.
Beukema W, de Pous P, Donaire D, Escoriza D, Bogaerts S, Toxopeus AG et al. (2010). Biogeography and contemporary climatic differentiation among Moroccan Salamandra algira. Biol J Linn Soc 101: 626–641.
Buckley D, Alcobendas M, García-París M, Wake MH (2007). Heterochrony, cannibalism, and the evolution of viviparity in Salamandra salamandra. Evol Dev 9: 105–115.
Bulut Z, McCormick CR, Gopurenko D, Williams RN, Bos DH, DeWoody JA (2009). Microsatellite mutation rates in the eastern tiger salamander (Ambystoma tigrinum tigrinum) differ 10-fold across loci. Genetica 136: 501–504.
Cabe PR, Page RB, Hanlon TJ, Aldrich ME, Connors L, Marsh DM (2007). Fine-scale population differentiation and gene flow in a terrestrial salamander (Plethodon cinereus) living in a continuous habitat. Heredity 98: 53–60.
Calado M (1994). Da Illha de Peniche In: Calado (eds). Peniche: Portugal.
Chakraborty R, Nei M (1977). Bottleneck effects on average heterozygosity and genetic distance with the stepwise mutation model. Evolution 31: 347–356.
Clegg SM, Degnan AM, Kikkawa J, Moritz Z, Estoup A, Owens IPF (2002). Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci USA 99: 8127–8132.
Cordero Rivera A, Velo-Antón G, Galán P (2007). Ecology of amphibians in small coastal Holocene islands: local adaptations and the effect of exotic tree plantations. Munibe 25: 94–103.
Cornuet JM, Luikart G (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Costas S, Muñoz Sobrino C, Alejo I, Pérez-Arlucea M (2009). Holocene evolution of a rock-bounded barrier-lagoon system, Cíes Islands, northwest Iberia. Earth Surf Proc Land 34: 1575–1586.
Dias JMA, Boski T, Rodrigues A, Magalhaes F (2000). Coast line evolution in Portugal since the Last Glacial Maximum until present - a synthesis. Mar Geol 170: 177–186.
Dopazo HJ, Alberch P (1994). Preliminary results on optional viviparity and intrauterine siblicide in Salamandra salamandra populations from northern Spain. Mertensiella 4: 125–138.
Dopazo HJ, Korenblum M (2000). Viviparity in Salamandra salamandra (Amphibia: Salamandridae): adaptation or exaptation? Herpetologica 56: 144–152.
Emerson BC (2002). Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol Ecol 11: 951–966.
Excoffier L, Laval G, Schneider S (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
García-París M, Alcobendas M, Buckley D, Wake DB (2003). Dispersal of viviparity across contact zones in Iberian populations of fire salamanders (Salamandra) inferred from discordance of genetic and morphological traits. Evolution 57: 129–143.
Garza JC, Williamson EG (2001). Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10: 305–318.
Gavrilets S, Vose A (2005). Dynamic patterns of adaptive radiation. Proc Natl Acad Sci USA 102: 18040–18045.
Gemmell NJ, Akiyama S (1996). An efficient method for the extraction of DNA from vertebrate tissues. Trends Genet 12: 338–339.
Goudet J, Raymond M, de Meeüs T, Rousset F (1996). Testing differentiation in diploid populations. Genetics 144: 1933–1940.
Grant PR (1998). Evolution on Islands. Oxford University Press: Oxford.
Greven H (1998). Survey of the oviduct of salamandrids with special reference to the viviparous species. J Exp Zool 282: 507–525.
Hartl DL, Clark AG (1997). Principles of Population Genetics. Sinauer Associates 3rd edition: Sunderland, MA.
Hedges SB, Hass CA, Maxson LR (1992). Caribbean biogeography: molecular evidence for dispersal in West Indian terrestrial vertebrates. Proc Natl Acad Sci USA 89: 1909–1913.
Hedrick PW, Cockerham CC (1986). Partial inbreeding: equilibrium heterozygosity and the heterozygosity paradox. Evolution 40: 856–861.
Heulin B, Osenegg K, Lebouvier M (1991). Timing of embryonic development and birth dates in oviparous and viviparous strains of Lacerta vivipara: testing the predictions of an evolutionary hypothesis. Acta Oecol 12: 517–528.
Heulin B, Osenegg-Leconte K, Michel D (1997). Demography of a bimodal reproductive species of lizard (Lacerta vivipara): survival and density characteristics of oviparous populations. Herpetologica 53: 432–444.
Hodges WL (2004). Evolution of viviparity in horned lizards (Phrynosoma): testing the cold-climate hypothesis. J Evol Biol 17: 1230–1237.
Hundertmark KJ, Van Daele LJ (2010). Founder effect and bottleneck signatures in an introduced, insular population of elk. Conserv Genet 11: 139–148.
Jakobsson M, Rosenberg NA (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806.
Kalinowski ST (2005). HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5: 187–189.
Kuhner MK (2006). LAMARC 2.00: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22: 6768–6770.
Lind MI, Ingvarsson PK, Johansson H, Hall D, Johansson F (2010). Gene flow and selection on phenotypic plasticity in an island system of Rana temporaria. Evolution 65: 684–697.
Losos JB, Ricklefs RE (2009). Adaptation and diversification on islands. Nature 457: 830–836.
Lovette IJ, Bermingham E, Ricklefs RE (2002). Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc Biol Soc 269: 37–42.
Luikart G, Cornuet JM (1998). Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12: 228–237.
MacArthur RH, Wilson EO (1967). The Theory of Island Biogeography. Princeton University Press: Princeton.
Martínez-Solano I, Lawson R (2009). Escape to Alcatraz: evolutionary history of slender salamanders (Batrachoseps) on the islands of San Francisco Bay. BMC Evol Bio 9: 38.
Mayr E (1954). Changes in genetic environment and evolution. In: Huxley J, Hardy AC, Ford EB (eds). Evolution as a Process. Allen and Unwin: London. pp 157–180.
Measey GJ, Vences M, Drewes RC, Chiari Y, Melo M, Bourles B (2007). Freshwater paths across the ocean: molecular phylogeny of the frog Ptychadena newtoni gives insights into amphibian colonization of oceanic islands. J Biogeogr 34: 7–20.
Meirmans PG (2006). Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60: 2399–2402.
Meirmans PG, Van Tienderen PH (2004). GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4: 792–794.
MeteoGalicia (2003). Anuario climatolóxico de Galicia. In Consellería de Medio Ambiente XdG (eds). Colección Tecnica Medio Ambiente. MeteoGalicia, Santiago de Compostela, Galicia, España.
Montori A, Herrero P (2004). Caudata. In: Ramos MA et al. (eds). Amphibia, Lissamphibia, vol. 24. García-París M, Montori A, Herrero P. Fauna Ibérica, Museo Nacional de Ciencias Naturales, CSIC, Madrid. pp 43–275.
Nei M, Maruyama T, Chakraborty R (1975). The bottleneck effect and genetic variability in populations. Evolution 29: 1–10.
Neill WT (1964). Viviparity in snakes: some ecological and zoogeographical considerations. Am Nat 98: 35–55.
Peakall R, Smouse PE (2006). GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Piry S, Luikart G, Cornuet JM (1999). BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90: 502–503.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Purrenhage JL, Niewiaroski PH, Moore FBG (2009). Population structure of spotted salamanders (Ambystoma maculatum) in a fragmented landscape. Mol Ecol 18: 235–247.
Qualls CP, Shine R (1998). Lerista bougainvillii, a case study for the evolution of viviparity in reptiles. J Evol Biol 11: 63–78.
Queller DC, Goodnight KF (1989). Estimating relatedness using genetic markers. Evolution 43: 258–275.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Shine R, Bull JJ (1979). Evolution of live bearing in lizards and snakes. Am Nat 113: 905–923.
Shine R (1985). The evolution of viviparity in reptiles: an ecological analysis. In: Gans C, Billet F (eds). Biology of the Reptilia. John Wiley & Sons: New York. pp 605–694.
Savage WK, Fremier AK, Shaffer HB (2010). Landscape genetics of alpine Sierra Nevada salamanders reveal extreme population subdivision in space and time. Mol Ecol 19: 3301–3314.
Spear SF, Peterson CR, Matocq M, Storfer A (2005). Landscape genetics of the blotched tiger salamander, Ambystoma tigrinum melanostictum. Mol Ecol 14: 2553–2564.
Steinfartz S, Küsters D, Tautz D (2004). Isolation and characterization of polymorphic tetranucleotide microsatellite loci in the Fire salamander Salamandra salamandra (Amphibia: Caudata). Mol Ecol Notes 4: 626–628.
Stockwell CA, Hendry AP, Kinnison MT (2003). Contemporary evolution meets conservation biology. Trends Ecol Evol 18: 94–101.
Templeton AR (1980). The theory of speciation via the founder principle. Genetics 94: 1011–1038.
Thompson JN (1998). Rapid evolution as an ecological process. Trends Ecol Evol 13: 329–332.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Veith M, Steinfartz S, Zardoya R, Seitz A, Meyer A (1998). A molecular phylogeny of ‘true’ salamanders (family Salamandridae) and the evolution of terrestriality of reproductive modes. J Zool Syst Evol Res. 36: 7–16.
Velo-Antón G, García-París M, Galán P, Cordero Rivera A (2007). The evolution of viviparity in Holocene islands: ecological adaptation vs phylogenetic descent along the transition from aquatic to terrestrial environments. J Zool Syst Evol Res 45: 345–352.
Vences M, Vieites DR, Glaw F, Brinkmann H, Kosuch J, Veith M et al. (2003). Multiple overseas dispersal in amphibians. Proc Soc Biol 270: 2435–2442.
Waddington CH (1942). Canalization of development and the inheritance of acquired characters. Nature 150: 563–565.
Wake MH (2004). Environmental effects, embryonization, and the evolution of viviparity. In: Hall BK, Pearson RD, Müller GB (eds). Environment, Development, and Evolution: Towards a Synthesis, vol. 4. MIT Press: Cambridge, MA. pp 151–170.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Weisrock DW, Papenfuss TJ, Macey JR, Litvinchuk SN, Polymeni R, Ugurtas IH et al. (2006). A molecular assessment of phylogenetic relationships and lineage accumulation rates within the family Salamandridae (Amphibia, Caudata). Mol Phyl Evol 41: 368–383.
Wourms JP (1981). Viviparity: the maternal–fetal relationship in fishes. Am Zool 21: 473–515.
Zamudio KR, Wieczorek AM (2007). Fine-scale spatial genetic structure and dispersal among spotted salamander (Ambystoma maculatum) breeding populations. Mol Ecol 16: 257–274.
Acknowledgements
Pedro Galán, María Casal Nantes and María Calviño helped to collect samples for this study. We thank the Zamudio Lab, David Buckley, Iñigo Martínez-Solano and three anonymous referees for constructive comments on earlier drafts of the manuscript. This research was funded by grants from the Spanish Ministry of Environment, and Organismo Autónomo Parques Nacionales (grant 072B/2002) to ACR. GVA was supported by a grant from the Galician Government (Consellería de Innovación, Industria e Comercio, Xunta de Galicia) and by a postdoctoral fellowship from the Spanish Ministerio de Ciencia e Innovación (Ref: 2008-0890). We thank the employees of the National Park for facilitating our trips to the islands, sometimes under difficult climatic conditions that complicated the access to San Martiño Island. Molecular data for this project were analyzed in the Evolutionary Genetics Core Facility and the Cornell Biological Resource Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Velo-Antón, G., Zamudio, K. & Cordero-Rivera, A. Genetic drift and rapid evolution of viviparity in insular fire salamanders (Salamandra salamandra). Heredity 108, 410–418 (2012). https://doi.org/10.1038/hdy.2011.91
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.91
Keywords
This article is cited by
-
Factors determining the dorsal coloration pattern of aposematic salamanders
Scientific Reports (2022)
-
Evolutionary effects of geographic and climatic isolation between Rhododendron tsusiophyllum populations on the Izu Islands and mainland Honshu of Japan
Heredity (2021)
-
Riverine barriers to gene flow in a salamander with both aquatic and terrestrial reproduction
Evolutionary Ecology (2021)
-
‘Mainland-island’ population structure of a terrestrial salamander in a forest-bocage landscape with little evidence for in situ ecological speciation
Scientific Reports (2020)
-
The evolution of pueriparity maintains multiple paternity in a polymorphic viviparous salamander
Scientific Reports (2020)