Abstract
We explored the origin of all-female broods resulting from male death in a Hokkaido population of Lymantria dispar through genetic crosses based on the earlier experiments done by Goldschmidt and by testing for the presence of endosymbionts that are known to cause male killing in some insect species. The mitochondrial DNA haplotypes of the all-female broods in Hokkaido were different from those of normal Hokkaido females and were the same as those widely distributed in Asia, including Tokyo (TK). Goldschmidt obtained all-female broods through backcrossing, that is, F1 females obtained by a cross between TK females (L. dispar japonica) and Hokkaido males (L. dispar praeterea) mated with Hokkaido males. He also obtained all-male broods by mating Hokkaido females with TK males. Goldschmidt inferred that female- and male-determining factors were weakest in the Hokkaido subspecies and stronger in the Honshu (TK) subspecies. According to his theory, the females of all-female broods mated with Honshu males should produce normal sex-ratio broods, whereas weaker Hokkaido sexes would be expected to disappear in F1 or F2 generations after crossing with the Honshu subspecies. We confirmed both of Goldschmidt's results: in the case of all-female broods mated with Honshu males, normal sex-ratio broods were produced, but we obtained only all-female broods in the Goldschmidt backcross and obtained an all-male brood in the F1 generation of a Hokkaido female crossed with a TK male. We found no endosymbionts in all-female broods by 4,′6-diamidino-2-phenylindole (DAPI) staining. Therefore, the all-female broods observed in L. dispar are caused by some incompatibilities between Honshu and Hokkaido subspecies.
Similar content being viewed by others
Introduction
Sexual reproduction is almost universal as a means of reproduction in a wide range of organisms and has the advantage of allowing gene exchanges to purge detrimental mutations and/or rapidly evolving parasites (Maynard Smith, 1978; Hamilton et al., 1990). Moreover, a two-sex system is generally used by most sexual organisms and has evolutionary advantages in numerous situations (Iwasa and Sasaki, 1987). Despite this, sex determination mechanisms are diverse and can differ even in closely related species (Peichel et al., 2004). In Drosophila melanogaster, sex is determined by the ratio of the chromosome number (X) to the number of the autosomal set (A) (Bridges, 1921). Animals with an X/A ratio of 1 become female, and those with an X/A ratio of 0.5 become male. In the housefly, Musca domestica, however, a male-determining factor (abbreviated as ‘M’) on the Y chromosome is critical for development into a male (Hediger et al., 1998). Without the ‘M’ factor, a female-determining factor (abbreviated as ‘F’) functions maternally, such that the animal develops as a female (Dübendorfer and Hediger, 1998).
Goldschmidt (1934) proposed the theory of sex determination of Lymantria dispar after studying hybrids among Japanese local races. In L. dispar, the heterogametic sex is female (ZW) and the homogametic sex is male (ZZ). Goldschmidt (1955) hypothesized that the female sex-determining factor, ‘F’, is a maternally inherited cytoplasmic factor and that the male sex-determining factor, ‘M’, is located on the Z chromosome (see Appendix; Figure 4). Goldschmidt (1934) suggested that different races of L. dispar possess female- and male-determining factors of different strengths. Within each race these factors are well balanced, but in inter-race crosses changes in the relative levels of these factors can generate intersex individuals or even cause sex reversal. Goldschmidt (1934) inferred that the female- and male-determining factors were weakest in Hokkaido subspecies (L. dispar praeterea) and stronger in Honshu (Tokyo (TK)) subspecies (L. dispar japonica). Goldschmidt (1934) obtained all-male broods from a cross between Hokkaido females and Honshu males. Inversely, Goldschmidt (1930) obtained all-female broods of L. dispar by backcrossing, that is, when F1 females resulting from a cross between Honshu females and Hokkaido males were mated with Hokkaido males (Appendix Table A1). The results of these crosses indicate that presumably weaker Hokkaido sexes disappear in the F1 or the backcrossed F2 generation when crossed with the other sex from the Honshu subspecies. The production of all-female or all-male broods is thus interesting in the context of sex determination in L. dispar.
Higashiura et al. (1999) found all-female broods owing to male death in L. dispar at Bibai (BB) in Hokkaido, Japan, and maternal inheritance of this trait. Higashiura et al. (1999) also observed six all-female broods out of 66 egg batches in the next generation in the same 0.2-ha forest, indicating the persistence of all-female broods. Clarke and Ford (1982) re-examined crosses between Hokkaido females and Honshu males and obtained all-male broods as Goldschmidt (1934) indicated. However, the characteristics of all-female broods obtained by the Goldschmidt backcross have not been re-examined.
If the Goldschmidt backcross indeed generates all-female broods in Hokkaido L. dispar, mitochondrial DNA (mtDNA) analysis of normal and all-female broods could reveal the maternal origin of the all-female broods. Bogdanowicz et al. (2000) described mtDNA variation among worldwide populations of L. dispar and resolved the haplotypes into four groups: (1) Okinawa, Japan, (2) Hokkaido, Japan, (3) Honshu and Kyushu, Japan, and mainland Asia, and (4) Europe, Tunisia, and North America. To test whether normal and all-female broods of Hokkaido L. dispar have different origins, we analysed mtDNA haplotypes in both broods. We also re-examined the Goldschmidt backcross that produces all-female broods at the backcrossed F2 generation. If the hypothesized weaker male sex of Hokkaido L. dispar indeed disappears in the Goldschmidt backcross, then females of the all-female broods mated with the hypothesized strong Honshu male should produce normal broods. In this study, we present the results of such crosses, which show that the origin of all-female broods is the hybridization between Honshu and Hokkaido subspecies.
At present, all-female broods are known to arise from selective male killing at egg or larval stages, particularly in arthropods (Majerus, 2003). Several microbial symbionts, including organisms in the genera Wolbachia, Spiroplasma, Rickettsia and Arsenophonus, are known to produce all-female broods in their host insects, as cytoplasmic elements are inherited only matrilineally (Hurst et al., 1997, 2003; Majerus, 2003). To test whether certain bacteria are present in all-female broods, diagnostic polymerase chain reaction (PCR) was carried out on the larvae of all-female broods of Hokkaido L. dispar using primers that amplify fragments of genes found in the above-mentioned bacteria and in eubacteria in general. As endosymbiont bacteria show up as bright flecks in the cytoplasm by 4′,6-diamidino-2-phenylindole (DAPI) staining, we also stained the eggs of all-female broods of Hokkaido L. dispar with DAPI to test whether certain symbionts were involved in the production of the all-female broods. Given that microsporidians are commonly found in lepidopteran insects (Agnew et al., 2003), we would expect DAPI staining to reveal both prokaryotic and eukaryotic endosymbionts in all-female broods.
The aims of this study were (i) to test whether microbial symbionts are involved in the production of all-female broods by diagnostic PCR and DAPI staining and (ii) to test whether all-female broods of L. dispar originate from hybridization between Honshu and Hokkaido subspecies by mtDNA analyses and the Goldschmidt backcross. This information will be useful in characterizing the molecular mechanisms of sex determination in L. dispar.
Materials and methods
Study species
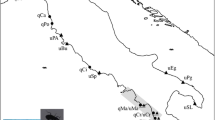
Inoue (1982) phenotypically classified L. dispar of Japan into five subspecies: L. dispar praeterea Kardakoff for Hokkaido populations, L. dispar japonica (Motschulsky) for the mainland (Honshu, Shikoku and Kyushu) populations (Figure 1) and three other island subspecies. All of these subspecies are univoltine. Schintlmeister (2004) treated the Hokkaido populations as L. dispar dispar, the same as mainland Asian and European populations. However, we adopted Inoue's classifications in this paper, because Hokkaido mtDNA haplotypes are quite different from Asian haplotypes, showing about 2% mtDNA sequence divergence (Bogdanowicz et al., 2000). Pogue and Schaefer (2007) reported that the distribution of L. dispar japonica is in Honshu, Shikoku and Kyushu, and extends north into parts of Hokkaido. They also treated Hokkaido populations as a distinct species, L. umbrosa (Butler). The type locality of the lectotype male for the revised species, L. umbrosa, is the most southwestern region of Hokkaido, or Hakodate (Figure 1). The distributions of the two species in Hokkaido are unclear. Therefore, we did not adopt the classifications described by Pogue and Schaefer (2007).
Map showing collection sites for egg masses of L. dispar used in this study. The arrow indicates North. Bibai (BB) (43°17′N, 141°51′E), Shikaoi (SK) (43°06′N, 142°57′E), Abira (AB) (42°45′N, 141°49′E), Niikappu (NP) (42°24′N, 142°14′E), Tomakomai (TM) (42°38′N, 141°37′E) and Asahikawa (AS) (43°49′N, 142°19′E) are in Hokkaido, Japan. Sannai (SN) (40°48′N, 140°40′E), Rifu (RF) (38°19′N, 140°56′E), Kawasaki (KW) (38°10′N, 140°40′E) and Tokyo (TK) (35°38′N, 139°23′E) are in Honshu, Japan. L. dispar praeterae is distributed in Hokkaido, Japan. L. dispar japonica is distributed in Honshu, Shikoku and Kyushu, Japan (Inoue, 1982). The Tsugaru Strait divides these two subspecies into their distributions. Hakodate, Hokkaido, is the type locality of L. umbrosa (Pogue and Schaefer, 2007).
In Hokkaido, larvae pupate in July and August. Adults emerge, mate and deposit eggs in August and September. As each female lays its full egg complement in only one mass containing 100–900 eggs, it is possible to recognize the properties of the original female by rearing eggs from individual masses. Eggs diapause during winter and hatch in the following May. Larvae feed on leaves of the Japanese larch Larix leptolepis, the Japanese birch Betula platyphylla and many other broad-leaved trees.
Sample collection and rearing of Hokkaido L. dispar
We collected egg masses at BB (Figure 1) and Shikaoi (SK) in Hokkaido, Japan. The collection sites were two Betula platyphylla forests (designated Betula-1 and Betula-2), Larix leptolepis forest at BB and one Betula platyphylla forest at SK. We collected and analysed seven egg masses from the Larix forest at BB in September 1999, six egg masses from Betula-1 forest at BB in October 1999 and 2001, 41 egg masses from Betula-2 forest at BB in October between 1999 and 2002, 11 egg masses at SK in November 2000 and 3 egg masses at Niikappu (NP) in December 2002. After collection, the egg masses were kept individually in a cold room at 0 °C until May of the following year. Then, about 100 eggs from each of the egg masses were placed in an incubator at 20 °C. After hatching, 20 first instar larvae from each of the egg masses were frozen in liquid nitrogen and stored at –80 °C for mtDNA analysis. The remaining larvae were reared to pupation on an artificial diet (product #F9642B, Bio-Serv, Frenchtown, NJ, USA). We counted unhatched eggs in some of the egg masses to obtain egg-hatch rates. Sex ratios among the pupae were recorded for each of the egg masses. As egg-hatch rates were <60% in all-female (male death) broods in the previous study (Higashiura et al., 1999), we regarded egg masses showing egg-hatch rates of 70% or more as normal sex-ratio broods without rearing.
PCR detection of bacterial symbionts
For PCR detection of bacterial symbionts, we used larvae hatched from three and two egg masses of all-female broods and normal broods, respectively, collected from Betula-2 forest at BB, Hokkaido, in October 1999. All of these broods were subjected to mtDNA sequencing as described in the next section. Five first instar larvae preserved in 99% ethanol from each of the egg masses were individually subjected to DNA extraction using a QIAamp Tissue Mini Kit (Qiagen, Hildeu, Germany). The purified DNA was eluted with 200 μl of TE buffer (10 mM Tris-HCl (pH 8.0), 0.1 mM ethylenediaminetetraacetic acid). For detection of potentially male-killing bacterial symbionts, diagnostic PCR analysis was conducted as described (Tsuchida et al., 2002). The following primers were used: ftsF and ftsR for the ftsZ gene of Wolbachia spp., 16SA1 and TKSSspR for the 16S rRNA gene of Spiroplasma spp., 16SA1 and Rick16SR for the 16S rRNA gene of Rickettsia spp., 16SA1 and Ars16SR for the 16S rRNA gene of Arsenophonus spp., 16SA1 and 16SB1 for the 16S rRNA gene of general eubacteria, and MtrA1 and MtrB1 for the mitochondrial rRNA gene of the host insect (Appendix Table A2). For positive control reactions, DNA samples from Callosobruchus chinensis (Coleoptera: Bruchidae) infected with Wolbachia sp., Drosophila melanogaster (Diptera: Drosophilidae) infected with Spiroplasma sp., Nephotettix cincticeps (Homoptera: Deltocephalidae) infected with Rickettsia sp. and Diaphorina citri (Homoptera: Psyllidae) infected with Arsenophonus sp. were also subjected to diagnostic PCR.
DAPI staining of embryos to detect endosymbionts
We collected the eggs of an all-female brood obtained by crossing a female of an all-female brood (44 females and 0 males) with a male of a normal brood (14 females and 16 males) collected at Abira (AB) in October 2005. As a control, we obtained eggs of normal broods by crossing individuals from two normal broods (a mother from a brood containing 18 females and 16 males, and a father from a brood containing 20 females and 17 males) collected at BB in October 2006. Eggs were collected from both all-female and normal sex-ratio broods at 3 days after oviposition. The collected eggs from each brood were treated with 50% (v/v) aqueous bleach solution for 30 s to 3 min. After washing with phosphate-buffered saline (PBS), eggs were fixed with Carnoy's fixative for 3 h in a siliconized tube at 4 °C. Subsequently, eggs were placed in PBS Tween for 15 min, and were treated with 50% aqueous bleach solution for 1 min before the surgical procedure. The chorion and vitelline membranes were removed with tungsten needles (cat. #500134, World Precision Instruments, Sarasota, FL, USA) under a microscope in PBS Tween solution. The dechorinated eggs were transferred to PBS Tween solution in another siliconized tube. A volume of 1 ml of DAPI solution (1 mg ml−1) was added and the mixture was incubated for 5 min. The stained eggs were observed under a fluorescence microscope using an ultraviolet filter.
As a positive control, Callosobruchus chinensis embryos infected with Wolbachia (strain jC) (Kondo et al., 2002) were stained using DAPI solution. We also evaluated C. chinensis without Wolbachia obtained by tetracycline treatment (Kondo et al., 2002). C. chinensis eggs were collected within 24 h after oviposition. The eggs were submerged in Carnoy's fixative with adzuki beans, and then detached from the beans. After removing the beans from the fixative, the eggs were fixed in a siliconized tube for 2 h at 4 °C. Eggs were rehydrated in PBS Tween solution for 15 min and stained with DAPI solution for 5 min. The observational method for stained C. chinensis eggs was the same as that used for L. dispar eggs.
mtDNA sequencing
In all, 5 or 10 frozen first instar larvae from each of the broods were ground with a pestle in a pre-chilled mortar and subjected to total DNA extraction using a QIAamp DNA Mini kit (Qiagen). In all, 1075 bp of mtDNA segment encoding the parts of three genes, cytochrome c oxidases I and II and nicotinamide adenine dinucleotide dehydrogenase subunit 1, was amplified by PCR from 2 ng of template DNA. The gene-specific primers have been described previously (Bogdanowicz et al., 2000) (Appendix Table A2). PCR (40 cycles of 94 °C for 0 s, 48 °C for 0 s and 72 °C for 15 s) was carried out using a Rapid Cycler (Idaho Technology, Salt Lake City, UT, USA). The amplified products were directly subjected to DNA sequencing using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit and ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The DNA sequences determined in this study have been deposited in the DDBJ, EMBL and GenBank nucleotide sequence databases under accession numbers AB244647–244657 and AB244665. Pairwise sequence-based distances among the studied mtDNA haplotypes (a total of 1075 base pairs across the three genes) were estimated using PAUP* 4.0b10 (Swofford, 2003) under the Hasegawa, Kishino and Yano (HKY) model (Hasegawa et al., 1985).
Crossing experiments
We collected two egg masses at Sannai (SN), Aomori in northern Honshu, Japan, in November 2001. The mtDNA haplotypes of these larvae were determined using the protocol described above. Hatched larvae were reared to adulthood and sexed in July 2002. The females were crossed with males from egg masses collected from Betula-2 at BB, Hokkaido, in October 2001. The offspring were reared to adulthood and sexed in July 2003. The F1 females were backcrossed with males with Hokkaido mtDNA haplotypes from egg masses of normal broods collected at NP, AB and Tomakomai (TM), Hokkaido, Japan in December 2002. The backcrossed F2 offspring were reared to adulthood and sexed in July 2004.
Females of all-female broods emerged from egg masses collected from Betula-2 at BB, Hokkaido, in October 2001. In July 2002, they were crossed either with (one) males from Honshu egg masses or (two) males from BB, Hokkaido; the latter cross served as a positive control. The Honshu egg masses were collected in November 2001 at Rifu (RF), Kawasaki (KW) and also at SN. Larvae from these crosses were reared to pupation and sexed in July 2003. An artificial diet (Bio-Serv; product #F9642B) was used for rearing.
A Hokkaido female that emerged from an egg mass collected at Asahikawa (AS) in April 2006 was crossed with a TK male from an egg mass collected at TK in October 2005. The offspring were reared and sexed in July 2007.
The exact P-value for the binomial proportion test (hereafter abbreviated as the binomial test) (SAS Institute, 1999) is used to analyse the egg hatch rate and sex ratio data.
Results
Detection of endosymbionts
Diagnostic PCR analysis of 15 larvae from three all-female broods of L. dispar detected no Wolbachia, Spiroplasma, Rickettsia, Arsenophonus or general eubacteria. These symbionts were also not detected in the 10 larvae from two normal sex-ratio broods. A mitochondrial gene of the host insect was successfully amplified from all of the samples, indicating that the DNA extraction was successful. Each positive control sample gave specific PCR products of the expected sizes for each bacterium and general eubacteria, confirming the specificity of the PCR (data not shown).
No differences were detected between L. dispar embryos of all-female and normal broods by DAPI staining (Figures 2a and b). However, in C. chinensis embryos infected with Wolbachia, we were able to detect a cytoplasm containing large amounts of Wolbachia (Figure 2c). In embryos without Wolbachia, which were obtained through tetracycline treatment, we found a single type of nucleus, lacking all apparent endosymbionts (Figure 2d).
Eggs stained with DAPI solution and viewed under a fluorescence microscope using an ultraviolet filter to detect endosymbionts. As DAPI stains nuclei exclusively, we observe a cytoplasm containing many parasitic bacteria. The length of each white bar represents 20 μm. (a) An embryo deposited by a female from an all-female brood of Lymantria dispar at 3 days after oviposition. (b) An embryo from a normal sex-ratio brood of L. dispar at three days after oviposition. (c) An embryo of Callosobruchus chinensis infected with Wolbachia (strain jC) (Kondo et al., 2002) within 24 h after oviposition. The arrow shows cytoplasm with abundant bacteria. (d) An embryo of C. chinensis without Wolbachia obtained by tetracycline treatment.
mtDNA sequences of all-female and normal sex-ratio broods of Hokkaido L. dispar
The mtDNA haplotypes of the all-female broods were found to be quite different from those of the normal sex-ratio broods (Figure 3). Pairwise mtDNA sequence divergence values from comparisons of the normal broods ranged from 0 to 0.0028 (substitution per site). The haplotype H1 in Figure 3 had been identified previously and was included in the Hokkaido haplotypes reported by Bogdanowicz et al. (2000). Nevertheless, the mtDNA haplotype (A1) of the all-female broods was the most common among those seen in Honshu (Japan), Kyushu (Japan) and mainland Asia (Bogdanowicz et al., 2000). Pairwise mtDNA sequence divergence values from comparisons of the all-female broods ranged from 0 to 0.0019. However, the sequence divergence between the normal and the all-female broods ranged from 0.016 to 0.019.
Variable sites of three genes, cytochrome c oxidases I and II (COI and COII), and nicotinamide adenine dinucleotide dehydrogenase subunit 1 (ND1), in the mtDNA of (the upper) all-female broods and (the lower) normal sex-ratio broods of Lymantria dispar, in Hokkaido, Japan. There are 17 fixed differences between all-female broods and normal sex ratio broods. These fixed different sites are shaded in grey. The 4–5-digit numbers in the upper column indicate the nucleotide position corresponding to the mtDNA sequence of Drosophila yakuba (Clary and Wolstenholme, 1985). ‘Number’ indicates the number of egg masses with the given haplotype among the egg masses collected at the locality. ‘A1’ is the same as the most common haplotype (28 out of 44 individuals) in Russia, China, Japan and Korea, and ‘H1’ is the same as the most common haplotype (3 out of 5 individuals) in Hokkaido, Japan (Bogdanowicz et al., 2000). Non-synonymous substitutions for A1 or H1 are boxed. BB, Bibai; NP, Niikappu; SK, Shikaoi (Figure 1).
The 18 all-female broods shown in Figure 3 contained no males at all. In one brood of the minimum rearing number shown in Figure 3, the sex ratio was 6:0 (females:males) (P(1:1)=0.03: the binomial test). In one brood of the maximum rearing number shown in Figure 3, the sex ratio was 29:0 (P(1:1) <0.001: the binomial test). The egg-hatch rates of the all-female broods were between 38 and 58%, indicating that all-female production was probably because of male death. The 50 normal sex-ratio broods in Figure 3 had more than 70% egg-hatch rate, or had a sex ratio of 1:1 (for example, the most skewed ratio of one normal brood in Figure 3, in which females:males=12:4; P(1:1)=0.08: the binomial test).
Crossing experiments
We used five normal sex-ratio broods of Honshu L. dispar in this study (Table 1). The mtDNA haplotype of all of these broods was A1, an Asian type distributed through Asia, including Japan, south of Hokkaido (Bogdanowicz et al., 2000). Of 15 broods of Hokkaido L. dispar used for crossing experiments in this study, 11 broods exhibited normal sex ratios near 1:1, whereas 4 broods consisted of all females. Egg-hatch rates were ⩾67% in the normal broods and ⩽56% in the all-female broods, indicating that all-female production was probably because of male death. In the Hokkaido L. dispar, the mtDNA haplotypes of the normal broods were H1 or H6, which are haplotypes that are found only in Hokkaido.
When the Honshu females and the Hokkaido males shown in Table 2, both of which came from broods with normal sex ratios, were crossed, the F1 offspring exhibited normal sex ratios. However, when F1 females were again crossed with Hokkaido males, the backcrossed F2 offspring were entirely female. Two series of independent experiments gave the same results (Table 2). These results are in perfect agreement with Goldschmidt's finding of all-female brood production in the hybridization crosses between Honshu females (L. dispar japonica) and Hokkaido males (L. dispar praeterea) (Goldschmidt, 1930; Appendix Table A1).
When Hokkaido females from all-female broods were mated with Hokkaido males, their offspring were all females in five replicates (Table 3a). These results confirm the result of Higashiura et al. (1999), indicating that the all-female broods produce all-female broods when females of these broods are mated with Hokkaido males.
When Hokkaido females of the all-female broods were mated with Honshu males, however, the offspring exhibited normal sex ratios of approximately 1:1 in five replicates (Table 3b).
We confirmed that an AS, Hokkaido, female mated with a TK, Honshu male produced an all-male brood. The egg hatch rate of the brood was 79%, which is significantly higher than 50% (P<0.0001: the binomial test; Table 4).
Discussion
Higashiura et al. (1999) found all-female broods in the Hokkaido population of L. dispar and showed that all-female broods mated with Hokkaido males produce all-female broods. Male killing caused by cytoplasmic bacterial symbionts has been reported in a variety of insects (Hurst et al., 1997, 2003; Majerus, 2003). Although we analysed the infection status of all-female and normal broods using PCR tests specific for Wolbachia, Rickettsia, Spiroplasma, Arsenophonus and general eubacteria, we found no bacteria in either all-female or normal broods of L. dispar larvae in Hokkaido. Moreover, we obtained cytological evidence showing that all-female broods of L. dispar do not have obvious prokaryotic or eukaryotic endosymbionts that might cause male killing, as we found no differences in DAPI staining between the embryos of the all-female brood and normal sex ratio. In contrast, the cytoplasm of Callosobruchus chinensis was stained by DAPI because of its high content of bacteria, or Wolbachia. These results suggest that male death in L. dispar may be caused by something other than endosymbionts, but do not exclude the possibility of viral influences. An RNA virus is reported to cause selective male-killing in the oriental tea tortrix, Homona magnanima (Nakanishi et al., 2008).
The mtDNA analysis of L. dispar performed in this study revealed the different origins of the all-female broods and normal sex-ratio broods found in the same populations in Hokkaido, Japan. The normal sex-ratio broods possessed Hokkaido mtDNA haplotypes, whereas the all-female broods had Asian (including Honshu) mtDNA haplotypes. Here we must consider the results of Goldschmidt's backcross (1930), that is, F1 females resulting from a cross between Honshu, Japan, females (L. dispar japonica) and Hokkaido males (L. dispar praeterea) mated with Hokkaido males, as an explanation for the all-female broods observed in Hokkaido.
We successfully reproduced Goldschmidt's result by backcrossing Honshu females with Hokkaido males, both of which were obtained from normal broods, and found that the backcrossed F2 progeny were entirely female (Figure 4; Appendix Equations (A1) and (A2)). On the other hand, when Hokkaido females from all-female broods were crossed with Honshu males, the offspring exhibited normal 1:1 sex ratios (Appendix Equation (A3)). The results of the crosses conform to the prediction made by Goldschmidt's theory. The all-female broods observed in a small fraction of Hokkaido females may have originated from hybridization between Honshu females and Hokkaido males. These all-female broods produce all-female broods as long as the females of these broods are mated with Hokkaido males.
Results of hybridization crosses in Lymantria dispar according to Goldschmidt's sex determination theory (Goldschmidt, 1934). Circles indicate the cytoplasm holding the female determinant factor (F). Large ovals indicate the Z chromosome holding the male determinant factor (M). ‘F’ and ‘M’ indicate sex determinant factors, not the chromosome. Small ovals indicate W chromosome. Open and grey symbols, circles and ovals, show weak and strong sex determinants, respectively. Female is ZW and male is ZZ. Honshu individuals have strong sex determinants indicated by ‘st’, and are represented by (Fst) Mst W for females, and (Fst) Mst Mst for males (parentheses indicate a cytoplasmic factor). Hokkaido individuals possess the weakest sex determinants, and are represented by (FHok) MHok W for females, and (FHok) MHok MHok for males. (a) All-female broods obtained by backcrossing, that is, F1 females of a cross between strong Honshu females (L. dispar japonica) and weak Hokkaido (Hok) males (L. dispar praeterea) mated with Hokkaido males. Lethality is indicated by the cross bar. (b) All-male broods obtained by crosses between weak Hokkaido females and strong Honshu males. The dashed cross bar indicates sex-reversed female, or male.
The origin of the Honshu females found in Hokkaido is not clear. Bogdanowicz et al. (2000) found that one of six analysed Hokkaido individuals carried an Asian mtDNA haplotype and proposed that this female may be ‘either a migrant from elsewhere in Asia or an individual of mixed ancestry’. We are currently investigating the distribution of mtDNA haplotypes throughout Hokkaido.
The reciprocal hybridization crosses between L. dispar individuals give the opposite results, producing all-male broods (Figure 4; Appendix Equation (A4); Goldschmidt, 1934). Clarke and Ford (1982) reassessed this cross, asking whether Hokkaido females mated with Honshu males would produce all-male progeny, including both sex-reversed males and normal males. Goldschmidt (1934) thought that F1 females of this cross had the weakest female and strong male sex determinants and would be transformed into males. Clarke and Ford (1982), however, found that all of the phenotypically male hybrids lacked the heterochromatic W chromosome found in normal females. They stated that ‘the excess of males in Goldschmidt's classic cross is due to the Haldane (1922) effect of genic imbalance and not to sex reversal’. We obtained an all-male brood by crossing a Hokkaido female with a TK male. The egg-hatching rate of the brood was significantly >50%. This implies that either incompatibilities may manifest before or after oviposition. However, Goldschmidt (1934) likely took the high hatching rate of all-male broods into consideration, as his work mentions the different survival rates of sex-reversed males and females. The survival rate and the presence of the heterochromatic W chromosome during larval stages of such all-male broods should be studied further, as Clarke and Ford (1982) may have evaluated the presence of the heterochromatic W chromosome in late-instar larvae, after the female larvae had died.
Hornett et al. (2006) proposed the existence of a suppressor gene against the male-killing trait caused by Wolbachia in the Southeast Asian and Polynesian nymphalid butterfly Hypolimnas bolina. Polynesian females infected with the bacteria exhibited the male-killing trait; however, Southeast Asian females produced 1:1 sex ratio progenies even when infected with the bacteria. Hornett et al. (2006) concluded that zygotic suppression is controlled at a single, genetically dominant locus because they obtained 1:1, 2:1 and 4:1 sex ratio progenies in the F1, backcrossed F2 and backcrossed F3 generations of Southeast Asian females (backcrossed with Polynesian males). However, in L. dispar, the presence of a suppressor gene only on the Z chromosomes of the Honshu and Asian backgrounds may be able to produce entirely all-female broods in the backcross, but we found no obvious symbionts.
Goldschmidt's theory of sex determination is based on the hypothesis that different races of L. dispar possess female- and male-determining factors of different strength (Goldschmidt, 1934). The balance within each race and the imbalance of sex-determining strength between different races can be considered to be the outcome of a coevolutionary ‘arms race’ between the sexes in a recent theory of sexual conflict (Arnqvist and Rowe, 2005). This theory is based on the presence of antagonistic interactions between females and males, for example, mating frequencies. At the species-specific level, these coevolutionary ‘arms races’ between the sexes are hidden by continuous adaptation and counter-adaptation, but they propel the evolution of the mating system in a group of insects (Arnqvist and Rowe, 2002). In L. dispar, the ‘arms races’ between the sexes are hidden in race-specific crosses, but the outcome of the conflict may occur in crosses between sexual races. The sex-determining genes reflect the outcome of this sexual conflict (Arnqvist and Rowe, 2005) in the manner indicated by Goldschmidt's theory of sex determination.
Goldschmidt (1934) pointed out that ‘an intersex is an individual which starts development with its original, gametic sex, but changes sex during development’. He regarded the timing of this change, or turning point, to be important and formulated the time law of intersexuality. Dietrich (2003) pointed out that ‘Goldschmidt was attempting a synthesis of evolutionary and developmental biology similar to the synthesis that has been emerging during the past 15 years (known as evolutionary developmental biology, or evo-devo)’. As shown in the results of the inter-race crosses in this paper, his theory should be re-evaluated, at least for L. dispar. Goldschmidt (1955) assumed that the ‘F’ factor on the W chromosome was the maternally inherited cytoplasmic factor. In insects such as the blowfly, hessian fly, fungus gnat and house fly, maternal factors are involved in female sex determination (Ullerich, 1984; Inoue and Hiroyoshi, 1986; Stuart and Hatchett, 1991; de Saint Phalle and Sullivan, 1996; Dübendorfer and Hediger, 1998; Sánchez and Perondini, 1999). On the basis of the results presented here, we suggest that the molecular mechanisms of male or female mortality may be revealed by crosses between different races with different sex strengths. Moreover, it is expected that the research investigating these molecular mechanisms will clarify the sexual determinants of L. dispar in the near future.
Accession codes
References
Agnew P, Becnel JJ, Ebert D, Michalakis Y (2003). Symbiosis of microsporidia and insects. In: Bourtzis K, Miller TA (eds). Insect Symbiosis. CRC Press: Boca Raton, FL. pp 145–163.
Arnqvist G, Rowe L (2002). Antagonistic coevolution between the sexes in a group of insects. Nature 415: 787–789.
Arnqvist G, Rowe L (2005). Sexual Conflict. Princeton University Press: Princeton, NJ.
Bogdanowicz SM, Schaefer PW, Harrison RG (2000). Mitochondrial DNA variation among worldwide populations of gypsy moths, Lymantria dispar. Mol Phylogenet Evol 15: 487–495.
Bridges CB (1921). Triploid intersexes in Drosophila melanogaster. Science 54: 252–254.
Clarke C, Ford EB (1982). Intersexuality in Lymantria dispar (L.). A further reassessment. Proc R Soc Lond B 214: 285–288.
Clary DO, Wolstenholme DR (1985). The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 22: 252–271.
Dietrich MR (2003). Richard Goldschmidt: hopeful monsters and other ‘heresies’. Nature Rev Genet 4: 68–74.
Dübendorfer A, Hediger M (1998). The female-determining gene F of the housefly, Musca domestica, acts maternally to regulate its own zygotic activity. Genetics 150: 221–226.
Fukatsu T, Shibao H, Nikoh N, Aoki S (2001). Genetically distinct populations in an Asian soldier-producing aphid, Pseudoregma bambucicola (Homoptera: Aphididae), identified by DNA fingerprinting and molecular phylogenetic analysis. Mol Phylogenet Evol 18: 423–433.
Goldschmidt R (1930). Untersuchungen über Intersexualität. V.. Z. Induct Abstl 56: 257–301.
Goldschmidt R (1934). Lymantria. Bibliogr Genet 11: 1–185.
Goldschmidt R (1955). Theoretical Genetics. University of California Press: Berkeley, CA.
Haldane JBS (1922). Sex-ratio and unisexual sterility in hybrid animals. J Genet 12: 101–109.
Hamilton WD, Axelrod R, Tanese R (1990). Sexual reproduction as an adaptation to resist parasites (review). Proc Natl Acad Sci USA 87: 3566–3573.
Hasegawa M, Kishino H, Yano T (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174.
Hediger M, Minet AD, Niessen M, Schmidt R, Hilfiker-Kleiner D, Çakir Ş et al. (1998). The male-determining activity on the Y chromosome of the housefly (Musca domestica L.) consists of separable elements. Genetics 150: 651–661.
Higashiura Y, Ishihara M, Schaefer PW (1999). Sex ratio distortion and severe inbreeding depression in the gypsy moth Lymantria dispar L. in Hokkaido, Japan. Heredity 83: 290–297.
Hornett EA, Charlat S, Duplouy AMR, Davies N, Roderick GK, Wedell N et al. (2006). Evolution of male-killer suppression in a natural population. PLoS Biol 4: 1643–1648.
Hurst GDD, Hurst LD, Majerus MEN (1997). Cytoplasmic sex-ratio distorters. In: O’Neill SL, Hoffmann AA, Werren JH (eds). Influential Passengers. Oxford University Press: Oxford. pp 125–154.
Hurst GDD, Jiggins FM, Majerus MEN (2003). Inherited microorganisms that selectively kill male hosts: the hidden players in insect evolution?. In: Bourtzis K, Miller TA (eds). Insect Symbiosis. CRC Press: Boca Raton, FL. pp 177–197.
Inoue H (1982). Lymantriidae. In: Inoue H, Sugi S, Kuroko H, Moriuti S, Kawabe A (eds). Moths of Japan, vol. 1. Text. Kodansha: Tokyo, Japan. pp 628–638.
Inoue H, Hiroyoshi T (1986). A maternal-effect sex-transformation mutation of the housefly, Musca domestica L. Genetics 112: 469–482.
Iwasa Y, Sasaki A (1987). Evolution of the number of sexes. Evolution 41: 49–65.
Kondo N, Ijichi N, Shimada M, Fukatsu T (2002). Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Mol Ecol 11: 167–180.
Majerus MEN (2003). Sex Wars: Genes, Bacteria, and Biased Sex Ratios. Princeton University Press: Princeton, NJ.
Maynard Smith J (1978). The Evolution of Sex. Cambridge University Press: Cambridge.
Mosbacher GC (1975). Sex specific cell differentiation in different types of intersexes of Lymantria dispar L. In: Reinboth R (ed.). Intersexuality in the Animal Kingdom. Springer: Berlin. pp 146–157.
Nakanishi K, Hoshino M, Nakai M, Kunimi Y (2008). Novel RNA sequences associated with late male killing in Homona magnanima. Proc R Soc B 275: 1249–1254.
Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J et al. (2004). The master sex-determination locus in three spine sticklebacks is on a nascent Y chromosome. Curr Biol 14: 1416–1424.
Pogue MG, Schaefer PW (2007). A Review of Selected Species of Lymantria Hübner [1819] (Lepidoptera: Noctuidae: Lymantriinae) from Subtropical and Temperate Regions of Asia, Including the Descriptions of Three New Species, Some Potentially Invasive to North America. US Department of Agriculture: Washington, DC.
de Saint Phalle B, Sullivan W (1996). Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early Sciara coprophila embryogenesis. Development 122: 3775–3784.
Sánchez L, Perondini ALP (1999). Sex determination in sciarid flies: a model for the control of differential X-chromosome elimination. J Theor Biol 197: 247–259.
SAS Institute (1999). The FREQ Procedure. SAS/STAT User’s Guide, Version 8. SAS Institute: Cary, NC, pp 1247–1362.
Schintlmeister A (2004). The taxonomy of the genus Lymantria Hübner, [1819] (Lepidoptera, Lymantriidae). Quadrifina 7: 1–248.
Stuart JJ, Hatchett JH (1991). Genetics of sex determination in the Hessian fly, Mayetiola destructor. J Heredity 82: 43–52.
Swofford D (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates: Sunderland, MA.
Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T (2002). Diversity and geographic distribution of secondary symbiotic bacteria in natural populations of the pea aphid Acyrthosiphon pisum. Mol Ecol 11: 2123–2135.
Ullerich FH (1984). Analysis of sex determination in the monogenic blowfly Chrysomya rufifacies by pole cell transplantation. Mol Gen Genet 193: 479–487.
Acknowledgements
We appreciate Jeff Oliver and three reviewers for their helpful comments to improve this paper. We thank Hideho Hara for collecting materials and Furumi Komai, Hywel Lloyd and Shin-ichi Akimoto for their comments on the paper. Masakazu Shimada and Natsuko Kondo kindly supplied us with Callosobruchus chinensis with and without Wolbachia infection. This work was financially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to Y Higashiura, H Yamagata and M Ishihara (No. 12440220 and 15370014).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Appendix
Appendix
A sex determination theory in Lymantria dispar by Goldschmidt (1934).
Goldschmidt (1934) proposed that the sex of L. dispar is determined by a quantitative balance between female and male sex determinants. He inferred that the strength of sex determinants is the same within each locality, but is different between localities. He divided Japanese L. dispar into eight races. He defined intersex as genetic females or males exhibiting intermediate phenotypes. The intersex does not occur in matings between insects in the same locality. However, it may occur when the balance of the strength of the sexual determinants collapses, as in matings between insects from different localities. Mosbacher (1975) confirmed the existence of the intersex by examining the shapes of forewing scales and antenna hairs in hybridization crosses of L. dispar. In his hypothesis (Goldschmidt, 1955), the female sex determinant (F) is located on the W chromosome, and each egg contains a W chromosome at least until meiosis, as the female is heterogametic, or ZW. Thus, each oocyte can express the female determinant on the W chromosome as a cytoplasmic factor, or a maternal factor. Goldschmidt (1955) assumed that the male sex determinant (M) is located on the Z chromosome and that it functions as a zygotic factor. In his notation, (F)MM denotes a ZZ male, two Ms from the two Z chromosomes and one F cytoplasmically inherited from the mother (parentheses indicate a cytoplasmic factor), whereas (F)MW denotes a ZW female, one M from the one Z chromosome and one F from the mother, with W indicating the presence of a W chromosome. (F)MM individuals develop into males because two Ms are stronger than one F, and (F)MW individuals become females because one M is weaker than one F. In hybridization, we must consider the different strength of sex determinants in individuals from each location. We focus on the Hokkaido and Honshu populations. The sex determinants of Hokkaido L. dispar (L. d. praeterea) for both sexes are inferred to be weakest among those investigated by Goldschmidt (1934), whereas those of Honshu L. dispar (L. d. japonica) are stronger. Other subspecies possess sex determinants of intermediate intensities; however, all of them are stronger than those of Hokkaido L. dispar. Given that the sex determinants of the Hokkaido L. dispar are denoted as FHok and MHok and those of the Honshu L. dispar as Fst and Mst, the cross between a Honshu female and a Hokkaido male can be represented as follows:

In the F1 generation, [(Fst) MHokW] individuals become females because the female determinant (Fst) is stronger than the male determinant (MHok); this outcome is compatible with the genetic sex (Figure 4). [(Fst) Mst MHok] individuals become males because Mst plus MHok is still stronger than Fst. The backcross between the F1 female and the Hokkaido male can be represented as follows:

In the backcrossed F2 generation, [(Fst) MHokW] individuals become females. However, it is hypothesized that [(Fst) MHok MHok] individuals (that is, extreme intersexual males) develop into females, because MHok plus MHok is weaker than Fst. Goldschmidt (1934) thought that females produced through sex reversal rarely survived. He stated that ‘it has to be added that the viability of the higher grades of intersexes is lowered and that therefore their number (of sex reversal females, that is, males) is smaller than expected’. In fact, he presents many examples of all-female broods produced by such weak males (Hokkaido males) mated with F1 females of a cross between many Japanese Honshu females and Hokkaido males (Appendix Table A1).
If all-female broods in Hokkaido occur in this manner, then the females of all-female broods mated with strong males or Honshu males may produce offspring in a normal sex ratio, according to Goldschmidt's theory (Goldschmidt, 1934), as follows:

These males are heterozygous normal males (Goldschmidt, 1934), and are the same as the F1 hybrid males shown in Equation (A1).
Reciprocal hybridization crosses of L. dispar give reverse results, producing all-male broods (Goldschmidt, 1934). In this case, Hokkaido females mated with Honshu males would produce all-male broods including both sex-reversed males and normal males (Figure 4). This cross can be shown as follows:

Goldschmidt (1934) thought that these F1 females had the weakest (FHok) female and strong male (Mst) sex determinants, and were transformed into males.
Rights and permissions
About this article
Cite this article
Higashiura, Y., Yamaguchi, H., Ishihara, M. et al. Male death resulting from hybridization between subspecies of the gypsy moth, Lymantria dispar. Heredity 106, 603–613 (2011). https://doi.org/10.1038/hdy.2010.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.92
Keywords
This article is cited by
-
Identification of Japanese Lymantria species (Lepidoptera: Lymantriidae) based on DNA sequences
Applied Entomology and Zoology (2018)
-
Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity
Scientific Reports (2017)
-
Defoliation by gypsy moths negatively affects the production of acorns by two Japanese oak species
Trees (2015)
-
Identification of Japanese Lymantria species (Lepidoptera: Lymantriidae) based on PCR–RFLP analysis of mitochondrial DNA
Applied Entomology and Zoology (2014)