Abstract
The genus Eigenmannia comprises several species groups that display a surprising variety of diploid chromosome numbers and sex-determining systems. In this study, hypotheses regarding phylogenetic relationships and karyotype evolution were investigated using a combination of molecular and cytogenetic methods. Phylogenetic relationships were analyzed for 11 cytotypes based on sequences from five mitochondrial DNA regions. Parsimony-based character mapping of sex chromosomes confirms previous suggestions of multiple origins of sex chromosomes. Molecular cytogenetic analyses involved chromosome painting using probes derived from whole sex chromosomes from two taxa that were hybridized to metaphases of their respective sister cytotypes. These analyses showed that a multiple XY system evolved recently (<7 mya) by fusion. Furthermore, one of the chromosomes that fused to form the neo-Y chromosome is fused independently to another chromosome in the sister cytotype. This may constitute an efficient post-mating barrier and might imply a direct function of sex chromosomes in the speciation processes in Eigenmannia. The other chromosomal sex-determination system investigated is shown to have differentiated by an accumulation of heterochromatin on the X chromosome. This has occurred in the past 0.6 my, and is the most recent chromosomal sex-determining system described to date. These results show that the evolution of sex-determining systems can proceed very rapidly.
Similar content being viewed by others
Introduction
Despite being largely conserved in birds and mammals, chromosomal sex-determining systems have evolved independently several times in the cold-blooded vertebrates (Devlin and Nagahama, 2002). Within fishes, a number of different sex-determining systems may be present even among closely related species (Peichel et al., 2004; Takehana et al., 2007), including those within Eigenmannia (Henning et al., 2008), and have recently been implicated in speciation as well (Kitano et al., 2009). The reason for this evolutionary lability is unknown, but may be related to the multiple redundant gene copies created by whole-genome duplication events (Meyer and Schartl, 1999) and duplications of single sex-determining genes (Nanda et al., 2002).
Traditional cytogenetic techniques have been able to identify heteromorphic sex chromosomes in only about 10% of the teleost species analyzed (Devlin and Nagahama, 2002). This number is expected to increase as the use of chromosome painting is increasingly becoming part of the fish cytogenetics toolkit (Phillips et al., 2001).
The glass knife weakly electric eels (Teleostei: Gymnotiformes: Eigenmannia) are distributed across most of South America and a part of Central America. They show several adaptations to the low visibility associated with riverine environments and their nocturnal lifestyle. These include morphological and behavioral adaptations; notably, the production of weak electric signals that are used for intraspecific recognition, communication and prey localization (Nelson, 2006). A number of gymnotiform species have been analyzed cytogenetically and shown a variety of diploid chromosome numbers, ranging from 2n=22 described in Apteronotus albifrons (Hinegard and Rosen, 1972) to 2n=54 in Gymnotus carapo (Foresti et al., 1984). The presence of differentiated sex chromosomes is not widespread across this order; however, a number of different sex-determining systems have been described (Almeida-Toledo and Foresti, 2001).
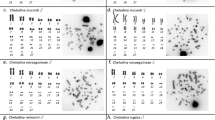
A number of different Eigenmannia cytotypes have been described from the major river basins in Brazil (Figure 1). Most of these populations are not distinguishable morphologically, owing in part to substantial intraspecific variability (Mago-Leccia, 1994). Species status has not been officially granted to several putative species, despite growing evidence for their reproductive isolation (Moysés et al., 2009). Two species of Eigenmannia with overlapping distributions in the Upper Paraná river basin present XY sex-determining systems—Eigenmannia virescens and the undescribed Eigenmannia sp.2 (Henning et al., 2008). E. virescens exhibits a karyotype of 2n=38XX:XY, whereas Eigenmannia sp.2 presents a karyotype consisting of 2n=31X1X2Y in males and 2n=32 X1X2X1X2 in females (Almeida-Toledo et al., 2000) (Figure 2).
(a) Giemsa-stained karyotype of a male specimen of E. virescens (2n=38XX:XY). Chromosome pairs were ordered by size. Pair 15 is sex determining. The heterochromatic block of the X chromosome is shown in the inset (bottom right). (b) Fluorescence in situ hybridization of a Y chromosome probe on a male metaphase of E. sp.2 showing the homology of a large metacentric to two acrocentric chromosomes. (c) Schematic representation of the sex chromosomes in E. sp.2 males and females.
Existing cytogenetic and population genetic evidence suggests that each of these species is sister to a taxon without differentiated sex chromosomes, with Eigenmannia sp.2 being sister to another undescribed species, Eigenmannia. sp.1 (Almeida-Toledo et al., 1988), and E. virescens 2n=38XX:XY being sister to E. virescens 2n=38 (Almeida-Toledo et al., 2001; Moysés et al., 2009). The distribution of these sister-taxa is mainly allopatric, with the exception of Eigenmannia sp.1 and sp.2, which have been found in sympatry in one locality (see Figure 1). The cytotypes with sex chromosomes are found in the Tietê River basin and the species without sex chromosomes occur in the Mogi-Guaçu River basin.
It was previously shown that these two heteromorphic sex chromosomes are not homologous and probably evolved independently (Henning et al., 2008). The processes proposed to account for the differentiation are fusion, in the branch leading to E. sp.2, and heterochromatinization, in the branch leading to E. virescens (Almeida-Toledo et al., 1988, 2001; Henning et al., 2008).
The objective of this study is to test these hypotheses and investigate the processes that have led to the evolution of differentiated sex chromosomes in Eigenmannia sp.2 and E. virescens 2n=38XX:XY. To achieve this, we identify (a) the sister species of each of these two species; (b) the chromosomes homologous to each sex chromosome in the sister species using chromosome painting and finally; (c) estimate the divergence to infer maximum dates of the two differentiated sex chromosomes.
We show that mtDNA supports the sister-species status of these two species pairs. The ortholog of the Y chromosome in E. sp.2 (whose differentiation is estimated at <6 mya) is fused to a different chromosome in E. sp.1 and this may have interesting evolutionary implications. The X chromosome in E. virescens was estimated to be <0.6 mya, making this the most recent differentiated sex chromosome described to date. The possible implications of these findings to the speciation processes in Eigenmannia and to the general theory of sex chromosome evolution are discussed.
Materials and methods
The experimental organism
For this study, the samples were sampled from the Upper Paraná, São Francisco and Amazon river basin. A total of 10 cytotypes were used for the phylogenetic analysis. Samples used for the cytogenetic analysis consisted of Eigenmannia sp.2; Eigenmannia sp.1 and E. virescens collected in small tributaries of the Tietê (23°05′S, 47°13′W) and Mogi-Guaçu (22°23′S, 47°25′W) rivers, respectively (Table 1, Figure 1).
The specimens were sent for taxonomic identification and classified into two species groups: E. virescens and E. trilineata (R Campos-da-Paz, personal communiction). The specimens analyzed in this work will be herein identified with relation to their cytotypes. Following previous publications (see Introduction section), the 2n=28, 2n=31X1X2Y/32X1X2X1X2, 2n=38 and 2n=38XX:XY cytotypes will be referred to as Eigenmannia sp.1, Eigenmannia sp.2, E. virescens and E. virescens (38XX:XY), respectively.
Molecular methods
Five regions of the mitochondrial genome were sequenced using standard methods and the following primer pairs Met-L4437+ND2-H4980 (Macey et al., 1997), ATP8.2-L8331+CO3.2-H9236 (Sivasundar et al., 2001), ND4-L11424+His-H12145 (Miya and Nishida, 1999), Cytb-L14841+Cytb-H15149 (Kocher et al., 1989) and L-Pro+H16498+3 (Borowsky and Mertz, 2001). GenBank accession numbers for these sequences are GQ228019-GQ228029 and GQ253014-GQ253056.
The ND4 sequence from the specimen with 2n=36 (Ec3Sf) could not be determined, and was treated as missing data. The sequences from these five regions were concatenated for the maximum parsimony (MP) and maximum likelihood (ML) phylogenetic analysis. Details of the data set used are given in the Supplementary Materials (Supplementary Table S1.1).
Alignment and model selection
Sequences obtained were aligned using ClustalW, as implemented in BioEdit (Hall, 1999) and corrected manually. Coding regions were translated for the alignment. The aligned data set contained 3016 characters. The nucleotide substitution model for the concatenated data set (ML analysis) and for each of the partitions (Bayesian analysis) was determined based on the hLRT and Akaike Information Criterion, respectively, as implemented in jModeltest 0.1.1(Posada, 2008).
Phylogenetic analysis
MP (Fitch, 1971) and ML analyses were conducted in PAUP v.4.0b10 (Swofford, 2002). MP analysis (heuristic search) was conducted with unweighted characters. Consistency and retention indices and bootstrap values were calculated using 1000 replications. Archolaemus blax (Gymnotiformes: Sternopygidae) was used as out-group for this analysis (see Alves-Gomes, 1999).
Partitioned Bayesian analysis was carried out using MrBayes (Ronquist and Huelsenbeck, 2003). Details on the partitioning schemes and selection methods are described in detail in the Supplementary File S1. Convergence was assessed by the average s.d. of split frequencies (printed on screen every 103 generations) and by visual inspection in Tracer v1.4.1 (available from http://www.beast.bio.ed.ac.uk/Tracer). The chosen partitioning scheme was then reanalyzed in MrBayes v.3.1 using 2 × 107 Markov Chain Monte Carlo generations with sampling every 100th generation; 25% of the samples were discarded as burnin and the remaining log file of each run was combined.
Cytogenetic character states (diploid chromosome number and presence of differentiated sex chromosomes) were mapped on to the Bayesian tree using MacClade v.4 (Maddison and Maddison, 2000).
Estimation of divergence times
Divergence times were estimated using BEAST v.1.5.2 (Drummond and Rambaut, 2007). Markov Chain Monte Carlo chains were run for 4 × 107 generations and sampled every 1000 generations. Tracer v.1.4.1 was used to calculate Bayes Factors and divergence estimates. Four different estimates were obtained using strict and relaxed clocks.
The relaxed clock analyses were performed using coding regions of ND2, ATPase 6 and Cytb partitioned by codon. The following taxa were used: E. virescens 2n=38, E. virescens 2n=38XY, E. sp.1, E. sp.2, zebrafish (gi∣8576324), carp (gi∣436882), stickleback (gi∣16267225), medaka (gi∣25005733), rainbow trout (gi∣82492328), salmon (gi∣3775976), fugu (gi∣23306916) and Tetraodon nigroviridis (gi∣86604632). Anguilla japonica (gi∣13429955) was used as an outgroup. Sequences were translated and aligned in Seaview using the Muscle algorithm (Galtier et al., 1996).
Maximum and minimum constraints on the uniform prior were provided for three nodes, following key calibration points indicated in the study by Benton et al. (2009) and available at http://www.timetree.org. An additional constraint of the Eigenmannia split was added to one of the analyses following Alves-Gomes (1999). Details of the partitioning schemes, nucleotide substitution models, rates and time constraints used are given in the Supplementary File S1 (Supplementary Table S1.2). The input file used in BEAST is available in Supplementary File S2.
Estimations using fixed rates were performed on the Eigenmannia data set. The Gymnotiform tRNA rate of 0.23%my described by Alves-Gomes (1999) was used to provide a conservative estimate (Mueller, 2006). A second estimate was performed using a teleost mtDNA rate of 0.7%my (Zardoya and Doadrio, 1999).
Cytogenetic analysis
Mitotic chromosomes were prepared as previously described (Almeida-Toledo et al., 2002). The sex chromosome probes were derived from the whole Y chromosome of Eigenmannia sp. 2 and the whole X chromosome of E. virescens (2n=38XX:XY), as described previously (Henning et al., 2008) using the methodology described by Weimer et al. (1999). Cross-species fluorescence in situ hybridization was performed as in Henning et al. (2008). For the analysis, chromosomes from the same metaphase were ordered by size. Size measurements were performed using ImageJ software (available from http://rsbweb.nih.gov/ij).
Probes for detection of telomeric sequences ttaggg(n) were produced using the method published by Ijdo et al. (1991b). Details of the protocol are given in the Supplementary File S3.
Simultaneous staining with 4′-6-diamidino-2-phenylindole and chromomycin A3 was performed by treating fresh slides with 100 μl of a solution containing 10 mM MgCl2 and 0.12 mg ml−1 chromomycin A3 for 2 h. Slides were then either mounted with Vectashield Mounting Medium with 4′-6-diamidino-2-phenylindole (Vector Labs, Burlingame, CA, USA) or treated for 10 min in a solution containing 0.2 ng ml−1 4′-6-diamidino-2-phenylindole and 2 × SSC.
Results
Phylogenetic analysis
MP analysis including 11 taxa and 3016 characters resulted in a single most parsimonious tree (1282 steps) (Figure 3a). The GTR+G model was determined to be the best evolutionary model for the concatenated data set under the hierarchical likelihood ratio test and was used in ML analysis. The best trees generated by ML and MP have similar topologies, and are combined in Figure 3a. The difference between the ML and MP trees is related to the position of 2n=34 and 2n=30 cytotypes (which are monophlyletic in the ML tree), and Eigenmannia sp. 2n=36 and Eigenmannia sp. 2n=38 (which are inverted in the ML tree). These differences concern the least supported nodes, and are indicated by asterisks in Figure 3a.
(a) Phylogenetic trees obtained by maximum parsimony (MP) and maximum likelihood (ML). MP and ML bootstrap values are shown, respectively, above and below the branches. Nodes not shared by both methods are labeled by *. (b) Partitioned Bayesian analysis. Posterior probabilities are shown above the branches. The species pairs used in the cross-painting experiments are highlighted and the taxa included in the E. trillineata species group (see Materials and methods section and Table 1) are labeled by **. Arrows in (b) indicate most parsimonious origin of sex chromosomes.
Partitioning scheme c (see Supplementary File S1) was chosen for the Bayesian analysis based on ln Bayes Factor comparison (Table 2 ). This scheme combined regions with similar evolutionary rates and models and resulted in a likelihood score indistinguishable from the most partitioned scheme, but with considerably less parameters. The Bayesian tree obtained is consistent with those from the MP and ML analyses. The differences are related to the lowest supported nodes mentioned above. In particular, the relationship of species 2n=36 VarA and 2n=36 VarB could not be resolved and is shown with a polytomy (Figure 3b).
All phylogenetic trees obtained using the several reconstruction methods revealed two lineages comprising the lower (2n=28–34) and higher (2n=36–38) diploid numbers, these lineages are hereafter referred to as L- and H-lineages, respectively (Figure 3). All trees show that the species that possess heteromorphic sex chromosomes are closest related to species lacking differentiated sex chromosomes. Specimens classified as members of the E. trillineata group were shown to be paraphyletic by all analysis (see Table 1).
Divergence time estimations
The estimates obtained using both relaxed and strict clocks are displayed in Table 3 . As expected, the strict clock based on tRNA rates always estimated more ancient divergent times. This analysis was included only to present a conservative estimate and will not be discussed further. The other estimates are consistent, especially in relation to the more recent splits. In short, the split of the two E. virescens species and the E. sp.1 and 2 species were estimated to be <0.55 and <7 ma, respectively, by all the remaining analyses. The mean split of the Eigenmannia H- and L-lineages is estimated to be between 27 and 36 ma. The tree obtained is available in the Supplementary Files (Supplementary Figure S1.2)
Cytogenetic analysis
Use of chromosome painting probes derived from microdissected chromosomes is known to produce relatively noisy signals, particularly, because of the presence of repetitive DNA that may severely bias degenerate oligonucleotide primed-PCR amplification, in which the number of template copies is small. In this study, only labeled euchromatic regions that appeared consistently in both homologous chromosomes were considered as an indication of homology.
The results of the hybridization of the Y-chromosome probe from Eigenmannia sp.2 on metaphases of Eigenmannia sp.1 revealed homology to a single arm of a large metacentric pair and to another pair of medium-sized acrocentric chromosomes (Figures 4a and b). Other consistent hybridization signals can also be seen on other chromosome pairs, notably, the centromeric regions of pair 2. These regions were shown to be heterochromatic by fluorochrome staining. In situ hybridization using telomeric probes did not reveal interstitial telomeric sequences in any of the specimens analyzed, despite the use of low stringency hybridization protocols (Supplementary File S3).
(a) Cross-species fluorescence in situ hybridization (FISH) using Y-chromosome probe obtained from Eigenmannia sp.2 hybridized to an Eigenmannia sp.1 metaphase. Pairs displaying consistent signals on euchromatic regions are indicated. (b) Chromosomes from the same metaphase ordered by size to aid interpretation of signals. (c) Cross-species FISH using X-chromosome probe obtained from E. virescens (2n=38XX:XY) hybridized to an E. virescens metaphase. Pairs displaying consistent signals on euchromatic regions are indicated. (d) Inverted 4′-6-diamidino-2-phenylindole image from same metaphase (to facilitate visualization of chromosome morphology). (e) Chromosome pairs from panel c ordered by size.
The X-specific probe derived from E. virescens (2n=38XX:XY) hybridized to a small pair of acrocentric chromosomes in E. virescens (Figures 4c–e). Some nonspecific signals were also located on euchromatic regions of other chromosome pairs; however, these signals were not considered to reflect homology as they were not shown in both of the homologs and are probably due to the presence of repetitive sequences in the probe.
MP mapping of the cytogenetic data (presence/absence of sex chromosomes and diploid number) on the phylogenetic tree generated by the partitioned Bayesian analysis could not determine the ancestral diploid number for each of the main lineages or the entire ingroup. Diploid numbers of 36 and 38 are equally parsimonious in the H-lineage. By assuming a hard polytomy (that is, multiple speciation events), 36 becomes the most parsimonious ancestral state. It is not clear, however, if such an assumption is warranted. The most parsimonious scenario of sex chromosome evolution is three independent origins (Figure 3b).
Discussion
The two chromosomal sex-determining systems investigated were found to be of very recent origin. The heterochromatinized XY system is the most recent system described to date (<0.6 my), and might provide additional insights for the study of patterns related to the early differentiation of sex chromosomes. Moreover, one of the chromosomes involved in the fusion event that gave rise to a multiple XY system is fused independently in the closest-related taxa and this probably constitutes an efficient post-mating barrier. The taxa that present heteromorphic sex chromosomes appear as sister taxa to species lacking differentiated sex chromosomes in all trees obtained. The evidence provided here through the use of chromosome painting suggests that ancestral chromosome pairs fused at least twice in the lineages leading to Eigenmannia sp.2 and Eigenmannia sp.1. In the former species, it has resulted in the evolution of a multiple XY system (Almeida-Toledo et al., 2000).
Overall, the data presented support the hypothesis of repeated evolution of differentiated sex chromosomes in Eigenmannia, involving fusions and heterochromatinization. These findings are summarized in Figure 5.
Schematic representation of the main findings and hypotheses generated by this study. (a) E. sp.1 and sp.2 diverged approximately 5 mya and fixed independent fusions involving the same chromosome (represented in blue). The fusion gave rise to a multiple XY sex-determination system in E. sp.2. The E. virescens species pair diverged more recently (<0.6 mya) and one of the species evolved heteromorphic sex chromosomes through the heterochromatinization of the distal region of the X chromosome. A hypothetical inversion might have led to this accumulation of heterochromatin. (b) Possible outcome of a hybridization event involving a E. sp.2 male and a E. sp.1 female. Half of the hybrids would inherit the E. sp.2 Y chromosome. This might lead to complications in the pairing of homologs during the hybrid meiosis. Possible sex ratio distortions and the outcome of the reciprocal cross are harder to deduce given the lack of knowledge of the sex-determination system of E. sp.1. Chromosomes derived from E. sp.1 and 2 are labeled with * and + respectively.
E. virescens species pair
Chromosome painting confirms the homology of the X chromosome of E. virescens (2n=38XX:XY) to a small acrocentric pair in E. virescens (Figures 4c–e). This species pair showed particularly high support (100% bootstrap in both MP and ML, and 1.0 Bayesian posterior) (Figure 3). Among all the species analyzed, this species pair was the only one to share inter-sequence simple repeats amplification patterns in a recent population genetic study (Moysés et al., 2009). Judging from the current allopatric distribution and divergence dates estimated (see Figure 1b and Table 3), it seems likely that suppression of recombination in the homologous pair of acrocentric chromosomes and accumulation of heterochromatin on the X chromosome occurred after a recent geographical separation.
It is unlikely that such a difference in heterochromatin content would result in any degree of post-mating isolation, although such a link has recently been suggested in Drosophila (Ferree and Barbash, 2009). It seems that the genetic differentiation is being maintained by geographic isolation alone, but possible negative effects on hybrid meiosis remains to be investigated. The accumulation of heterochromatin in one chromosome of a pair (in this case, the X chromosome) implies that there is already restricted recombination, and the most common explanation is the presence of inversions (Charlesworth, 1991).
The observation that the X chromosome of this system is heterochromatinized seems puzzling. Recombination restriction is more drastic in the regions surrounding the sex-determination locus of the Y chromosome and is, therefore, expected to lead to a degeneration and a higher degree of repetitive DNA expansion on the Y chromosome (Charlesworth, 1991). However, the accumulation of heterochromatin in the X chromosome might be simply related to the recent age of this sex-determination system. It is known that levels of heterochromatin can vary substantially, and, in fact, variation in the size of this heterochromatic block has already been observed in a natural population of E. virescens (Almeida-Toledo et al., 2001). Recently, four Eigenmannia karyotypes bearing ZW sex chromosomes were described from the Amazon basin (Silva et al., 2009). Based conventional cytogenetic techniques, the authors suggest independent origins and that the ancestral sex chromosome pair is homologous to the Eigenmannia virescens XY system studied here (see Figure 5 in Silva et al., 2009). Future studies combining chromosome painting and phylogenetics are needed to test these hypotheses.
Eigenmanna sp.1 and sp.2
Eigenmannia sp.1 and Eigenmannia sp.2 also appear as sister taxa, and the fusion event leading to the evolution of the neo-Y chromosome is also recent (<6 ma). The similarity between the karyotypes of these two taxa had already been observed using R banding and supports the sister-species status of this species pair (Almeida-Toledo et al., 1988). The fact that one of the chromosomes homologous to the Y chromosome is fused to two different chromosomes in this species pair may have interesting evolutionary implications (Figure 5b). This is likely to result in a significant level of infertility in potential F1 hybrids because of abnormal meiotic segregation (Wallace et al., 2002). This effect would likely be compounded if the chromosome happened to harbor sex-determining genes, given the disproportionate importance of sex chromosomes in the evolution of post-mating isolation mechanisms (Presgraves, 2008; Kitano et al., 2009).
In contrast to the E. virescens species pair, the karyotypic differences probably constitute a post-mating reproductive isolation mechanism, and it is likely that these species would not hybridize significantly in the absence of geographic isolation. In fact, these two cytotypes have been found in sympatry (CB Moysés, personal observation). An exciting possibility is that stronger mating discrimination has evolved between the sympatric populations in the process known as reinforcement of reproductive isolation. In this case, presumably, the increase in levels of premating isolation might involve the reproductive character displacement of electric signals associated with species recognition (Feulner et al., 2009). Similarly, the sympatric distributions of Eigenmannia sp.1 and E. virescens; and Eigenmannia sp.2 and E. virescens (2n=38XX:XY) are also an indication of significant levels of reproductive isolation.
No evidence of interstitial telomeric sequences was found in Eigenmannia sp1 or Eigenmannia sp.2, despite evidence for the occurrence of multiple fusion events. Explanations for this pattern may involve the loss of one centromere and surrounding sequences in the case of a centric fusion and the rapid evolution of the pericentromeric heterochromatin. Losses of regions surrounding the fusion site have been described in several groups (Nanda et al., 1995; Perez et al., 1999). Alternatively, this might reflect insufficient resolution in our assay—it has been shown, for example, that the human chromosome 2 harbors ancient telomeric sequences in the vicinity of the centromeric region (2q13), but that these can only be detected with higher resolution techniques (Ijdo et al., 1991a). Either way, the hypothesis that the Y chromosome originated by fusion is further supported by the observation that the Y chromosome is homologous to two different autosomal pairs in E. virescens, an outgroup to this species pair (Henning et al., 2008). Thus, the most parsimonious ancestral state is that of two pairs.
Concluding remarks
Theoretical models have shown that the rate of fixation of a chromosomal rearrangement is proportional to the mutation rate and selection, and is inversely proportional to effective population size (Lande, 1979). Considering the likely negative initial fitness effects of multiple rearrangements, it is possible that the current pattern was aided by random processes of deme extinction followed by recolonization by a few individuals. Indeed, the failure to collect specimens in previously sampled localities (LF de Almeida-Toledo and F Henning, personal observation) suggests that these populations are highly unstable. If a link between fixation of these chromosome rearrangements and speciation where upheld by future studies, it might provide additional support to the mayrian view of speciation occurring mainly through founder events with no direct link to ecological factors and natural selection (Mayr, 1942). It should be noted, however, that ecological differences between these species remain to be investigated. Sex-specific ecological strategies also remain unknown, and might be important for the evolution of differentiated sex chromosomes.
In conclusion, the Eigenmannia species group has provided several independent examples of the rapid evolution of sex chromosomes and of chromosomal rearrangements (Almeida-Toledo et al., 2000, 2001, 2002; Almeida-Toledo and Foresti, 2001; Henning et al., 2008). The combined approach of phylogenetic and molecular cytogenetic methods used in this study highlights the occurrence of a series of chromosomal rearrangements that gave raise to the contemporary pattern of karyotypic variation in Eigenmannia and may have significant implications for the maintenance, and possibly the origin of reproductive isolation in this interesting species complex.
Accession codes
References
Almeida-Toledo L, Viegas-Péquignot E, Foresti F, Toledo Filho S, Dutrillaux B (1988). BrdU replication patterns demonstrating chromosome homoeologies in two fish species, genus Eigenmannia. Cytogenet Cell Genet 48: 117–120.
Almeida-Toledo LF, Daniel-Silva MFZ, Moyses CB, Fonteles SBA, Lopes CE, Akama A et al. (2002). Chromosome evolution in fish: sex chromosome variability in Eigenmannia virescens (Gymnotiformes:Sternopygidae). Cytogenet Genome Res 99: 164–169.
Almeida-Toledo LF, Foresti F (2001). Morphologically differentiated sex chromosomes in neotropical freshwater fish. Genetica 111: 91–100.
Almeida-Toledo LF, Foresti F, Daniel MFZ, Toledo SA (2000). Sex chromosome evolution in fish: the formation of the neo-Y chromosome in Eigenmannia (Gymnotiformes). Chromosoma 109: 197–200.
Almeida-Toledo LF, Foresti F, Pequignot EV, Daniel-Silva MF (2001). XX:XY sex chromosome system with X heterochromatinization: an early stage of sex chromosome differentiation in the Neotropic electric eel Eigenmannia virescens. Cytogenet Cell Genet 95: 73–78.
Alves-Gomes JA (1999). Systematic biology of gymnotiform and mormyriform electric fishes: Phylogenetic relationships, molecular clocks and rates of evolution in the mitochondrial rRNA genes. J Exp Biol 202: 1167–1183.
Benton M, Donoghue P, Asher R (2009). Calibrating and constraining molecular clocks. In: Hedges S and Kumar S (eds). The Timetree of Life. Oxford University Press: New York, pp 35–86.
Borowsky RB, Mertz L (2001). Genetic differentiation among populations of the cave fish Schistura oedipus (Cypriniformes: Balitoridae). Environ Biol Fishes 62: 225–231.
Charlesworth B (1991). The evolution of sex chromosomes. Science 251: 1030–1033.
Devlin RH, Nagahama Y (2002). Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364.
Drummond AJ, Rambaut A (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214.
Feulner PGD, Plath M, Engelmann J, Kirschbaum F, Tiedemann R (2009). Electrifying love: electric fish use species-specific discharge for mate recognition. Biol Lett 5: 225–228.
Ferree PM, Barbash DA (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol 7: e1000234.
Fitch WM (1971). Toward defining course of evolution: minimum change for a specific tree topology. Syst Zool 20: 406–416.
Foresti F, Almeida-Toledo LF, Toledo-Filho SA (1984). Chromosome studies in Gymnotus carapo and Gymnotus sp. (Pisces, Gymnotidae). Caryologia 37: 141–146.
Galtier N, Gouy M, Gautier C (1996). SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548.
Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98.
Henning F, Trifonov V, Ferguson-Smith MA, Almeida-Toledo LF (2008). Non-homologous sex chromosomes in two species of the genus Eigenmannia (Teleostei: Gymnotiformes). Cytogenet Genome Res 121: 55–58.
Hinegard R, Rosen DE (1972). Cellular DNA content and evolution of teleostean fishes. Am Nat 106: 621–644.
Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA (1991a). Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA 88: 9051–9055.
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991b). Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19: 4780.
Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF et al. (2009). A role for a neo-sex chromosome in stickleback speciation. Nature 461: 1079–1083.
Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX et al. (1989). Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86: 6196–6200.
Lande R (1979). Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 33: 234–251.
Macey JR, Larson A, Ananjeva NB, Fang ZL, Papenfuss TJ (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol 14: 91–104.
Maddison DR, Maddison WP (2000). MacClade 4: Analysis of phylogeny and character evolution. Sinauer: Sunderland, MA.
Mago-Leccia F (1994). Electric Fishes of the Continental Waters of America. Fundacion para el Desarrollo de las Ciencias Fisicas, Matematicas y Naturales: Caracas, 225pp.
Mayr E (1942). Systematics and the Origin of Species. Columbia University Press: New York, 334pp.
Meyer A, Schartl M (1999). Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 11: 699–704.
Miya M, Nishida M (1999). Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol 1: 416–426.
Moysés C, Daniel-Silva M, Lopes CE, Almeida-Toledo L (2009). Cytotype-specific ISSR profiles and karyotypes in the neotropical genus Eigenmannia (Teleostei: Gymnotiformes). Genetica 138: 179–189.
Mueller RL (2006). Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst Biol 55: 289–300.
Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A et al. (2002). A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA 99: 11778–11783.
Nanda I, Schneider-Rasp S, Winking H, Schmid M (1995). Loss of telomeric sites in the chromosomes of Mus musculus domesticus (Rodentia: Muridae) during Robertsonian rearrangements. Chromosome Res 3: 399–409.
Nelson JS (2006). Fishes of the World, 4th edn. Wiley: Hoboken, NJ, 601pp.
Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J et al. (2004). The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol 14: 1416–1424.
Perez J, Moran P, Garcia-Vazquez E (1999). Cloning and physical mapping of Atlantic salmon (Salmo salar L.) telomeric sequences. Heredity 82: 409–414.
Phillips RB, Konkol NR, Reed KM, Stein JD (2001). Chromosome painting supports lack of homology among sex chromosomes in Oncorhynchus, Salmo, and Salvelinus (Salmonidae). Genetica 111: 119–123.
Posada D (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256.
Presgraves DC (2008). Sex chromosomes and speciation in Drosophila. Trends Genet 24: 336–343.
Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Sivasundar A, Bermingham E, Orti G (2001). Population structure and biogeography of migratory freshwater fishes (Prochilodus : Characiformes) in major South American rivers. Mol Ecol 10: 407–417.
Swofford D (2002). PAUP* v. 4 Phylogenetic analysis using parsimony (* and other methods). Sinauer Associates Sunderland, MA.
Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M (2007). Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116: 463–470.
Wallace BMN, Searle JB, Everett CA (2002). The effect of multiple simple Robertsonian heterozygosity on chromosome pairing and fertility of wild-stock house mice (Mus musculus domesticus). Cytogenet Genome Res 96: 276–286.
Weimer J, Kiechle M, Senger G, Wiedemann U, Ovens-Raeder A, Schuierer S et al. (1999). An easy and reliable procedure of microdissection technique for the analysis of chromosomal breakpoints and marker chromosomes. Chromosome Res 7: 355–362.
Zardoya R, Doadrio I (1999). Molecular evidence on the evolutionary and biogeographical patterns of European cyprinids. J Mol Evol 49: 227–237.
Acknowledgements
This work was funded by grants from the State of São Paulo Research Foundation (FAPESP) to CBM (02/09717-0) and FH (05/51629), and grants from National Council for Research and Development (CNPq) to LFAT. Further support was provided by a grant from the Brazilian National Counsel of Scientific and Technologic Development (CNPq)/German Academic Exchange Program (DAAD) to FH and grant from the Deutsche Forschungsgemeinschaft (DFG) to AM. We thank Dr Ricardo Campos-Paz for specimen identification; Carlos Eduardo Lopes, Dr Alberto Akama, Dr Flora Fernandes, Riviane Garcez, Sabrina Baroni and Dr Soraia Barreto for support during collection of samples. We also thank Chris Baker for the critical reading of an earlier version of this article, Dr Kathryn Elmer and two anonymous reviewers for helpful suggestions and Tereza Manousaki for writing the script for combining the Mr Bayes log files. Professor Malcolm Ferguson-Smith, Professor Eduardo Gorab and Dr Vladimir Trifonov for providing assistance and allowing the use of facilities to develop the chromosome probes used.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Rights and permissions
About this article
Cite this article
Henning, F., Moysés, C., Calcagnotto, D. et al. Independent fusions and recent origins of sex chromosomes in the evolution and diversification of glass knife fishes (Eigenmannia). Heredity 106, 391–400 (2011). https://doi.org/10.1038/hdy.2010.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.82
Keywords
This article is cited by
-
Sex-Specific Genomic Region Identification and Molecular Sex Marker Development of Rock Bream (Oplegnathus fasciatus)
Marine Biotechnology (2022)
-
Sex chromosome evolution in frogs—helpful insights from chromosome painting in the genus Engystomops
Heredity (2021)
-
Against the mainstream: exceptional evolutionary stability of ZW sex chromosomes across the fish families Triportheidae and Gasteropelecidae (Teleostei: Characiformes)
Chromosome Research (2021)
-
Evidence for a Robertsonian fusion in Solea senegalensis (Kaup, 1858) revealed by zoo-FISH and comparative genome analysis
BMC Genomics (2018)
-
The transcriptomic signature of different sexes in two protogynous hermaphrodites: Insights into the molecular network underlying sex phenotype in fish
Scientific Reports (2018)