Abstract
Ejaculates comprise multiple and potentially interacting traits that determine male fertility and sperm competitiveness. Consequently, selection on these traits is likely to be intense, but the efficacy of selection will depend critically on patterns of genetic variation and covariation underlying their expression. In this study, I provide a prospective quantitative genetic analysis of ejaculate traits in the guppy Poecilia reticulata, a highly promiscuous livebearing fish. I used a standard paternal half-sibling breeding design to characterize patterns of genetic (co)variation in components of sperm length and in vitro sperm performance. All traits exhibited high levels of phenotypic and additive genetic variation, and in several cases, patterns of genetic variation was consistent with Y-linkage. There were also highly significant negative genetic correlations between the various measures of sperm length and sperm performance. In particular, the length of the sperm's midpiece was strongly, negatively and genetically correlated with sperm's swimming velocity—an important determinant of sperm competitiveness in this and other species. Other components of sperm length, including the flagellum and head, were independently and negatively genetically correlated with the proportion of live sperm in the ejaculate (sperm viability). Whether these relationships represent evolutionary trade-offs depends on the precise relationships between these traits and competitive fertilization rates, which have yet to be fully resolved in this (and indeed most) species. Nevertheless, these prospective analyses point to potential constraints on ejaculate evolution and may explain the high level of phenotypic variability in ejaculate traits in this species.
Similar content being viewed by others
Introduction
Sperm competition, the contest between ejaculates to fertilize eggs (Parker, 1970), is a common and powerful evolutionary force generating intense selection on ejaculates (Birkhead and Møller, 1998; Pizzari and Parker, 2009). Although sperm competition theory largely focuses on variation in traits linked to sperm production, including sperm numbers and testes size, there is increasing evidence that differences in the relative ‘quality’ of competing ejaculates can also generate variation in competitive fertilization rates (Snook, 2005). Such traits include sperm swimming velocity, sperm viability (the proportion of live sperm in a male's ejaculate) and sperm length—all of which have been shown to predict competitive fertilization success or fertility in several taxa. For example, differences in sperm swimming velocity between competing ejaculates is the primary determinant of competitive fertilization success in some birds (Birkhead et al., 1999; Denk et al., 2005) and fishes (Gage et al., 2004; Liljedal et al., 2008; Skjaeraasen et al., 2009), and is associated with fertility (non-competitive fertilization rates) in mammals (Gomendio and Roldan, 2008). Similarly, sperm viability predicts competitive fertilization success in insects (García-González and Simmons, 2005) and is associated with the strength of selection from sperm competition across insect taxa (Hunter and Birkhead, 2002). The relationships between sperm length and competitive fertilization success are more varied than for velocity and viability. In some species, sperm length is positively associated with sperm's swimming velocity (for example, Mossman et al., 2009; Fitzpatrick et al., 2010), which may explain macroevolutionary patterns of sperm evolution, which suggest that species with relatively high levels of sperm competition produce longer sperm (Fitzpatrick et al., 2009; Tourmente et al., 2009). Despite these latter observations, however, the relationships between sperm length and sperm performance at the intraspecific level can be positive and negative (see Humphries et al., 2008), and some studies have shown that relatively short sperm can have a competitive advantage in sperm competition (Gage and Morrow, 2003; García-González and Simmons, 2007; Firman and Simmons, 2008).
An implicit assumption in all discussions of sperm evolution is that ejaculate traits exhibit sufficient additive genetic variation to facilitate evolutionary responses to selection, and that patterns of genetic covariation among ejaculate traits do not constrain these processes (see review by Evans and Simmons, 2008). Thus, genetic approaches for studying sperm evolution are essential, and especially those that focus on multiple traits in order to shed light on their patterns of genetic covariance, and therefore the potential for genetic correlations to either facilitate or impede multivariate selection on ejaculate traits (for example, Simmons and Kotiaho, 2002; Moore et al., 2004; Birkhead et al., 2005; Snook et al., 2010). Although an increasing (but still small) body of literature suggests that individual ejaculate traits can exhibit very high levels of additive genetic variation (reviewed by Simmons and Moore, 2009), there is also accumulating evidence that trade-offs between various ejaculate traits may constrain their evolvability. For example, in the cockroach, Nauphoeta cinerea, sperm viability is negatively genetically correlated with both testes mass and sperm numbers (Moore et al., 2004), whereas in the zebra finch, Taeniopygia guttata, midpiece size is negatively genetically correlated with the length of the sperm's head and flagellum (Birkhead et al., 2005). Both studies therefore highlight the potential for trade-offs to impede evolutionary responses to selection on individual ejaculate traits.
In the present study, I provide a prospective analysis of quantitative genetic variation and covariation in ejaculate traits in the guppy, Poecilia reticulata, a livebearing freshwater poeciliid fish with a highly promiscuous mating system (Magurran, 2005). The study complements recent quantitative genetic analyses of pre and postcopulatory sexually-selected traits in this species in which two measures of in vitro sperm performance—sperm swimming velocity (average path velocity; VAP), and the proportion of live sperm per ejaculate (sperm viability)—were shown to be negatively genetically associated with male sexual behaviour and body ornamentation (Evans, 2010). The present study extends these analyses by exploring patterns of genetic covariation among ejaculate traits, adding to just handful of studies that have addressed this topic in other species (reviewed by Simmons and Moore, 2009).
Ejaculate traits and sperm competition in the guppy
Although, we currently lack a complete understanding of how individual ejaculate traits function during sperm competition in guppies, there is some evidence that several traits are important in this regard. For example, recent data confirm that sperm swimming velocity is the primary determinant of competitive fertilization success in this species (C Boschetto, C Gasparini and A Pilastro; unpublished data), which follows a general pattern across fishes and other taxa (see above). Furthermore, the size of the sperm's midpiece has been shown to be significantly phenotypically positively associated with sperm swimming velocity in guppies (Pitcher et al., 2007; Skinner and Watt, 2007), suggesting that this trait may also have a functional role in sperm competition. Indeed, a recent intraspecific comparative study on natural guppy populations in Trinidad revealed that males inhabiting populations characterized by high levels of female multiple mating (and therefore, presumably high levels of sperm competition) possess significantly faster swimming sperm with longer midpieces, than males from streams where female multiple mating is less frequent (Elgee et al., 2010). Taken together, these studies suggest that selection imposed by sperm competition will favour males producing sperm with enhanced sperm swimming velocity and longer midpieces. The present analysis therefore focuses on patterns of genetic (co)variation in these and other prospective measures of sperm quality to determine the extent to which genetic correlations among these traits may either impede or facilitate ejaculate evolution.
Materials and methods
Study population and breeding design
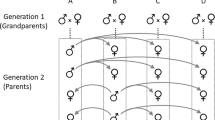
The fish used in this study were descendents of guppies captured in 2006 from the Alligator Creek River in Queensland, Australia. Guppies were introduced to this location approximately 100 years ago, probably from Guyana, West Indies (Lindholm et al., 2005). A standard nested paternal half-sibling/full-sibling breeding design was established by artificially inseminating the sperm from each of 40 males (sires) into five females (dams; 200 in total). Artificial insemination was used to minimize differential maternal effects, which are known in guppies (Pilastro et al., 2004) and may inflate sire estimates of additive genetic variation (for example, Kotiaho et al., 2003). Following inseminations, females were maintained individually in 3 l containers until they produced their first brood, which were removed from their natal tank and maintained in family groups until they were 13 weeks old. At this time, male offspring were isolated and placed individually into 2 l (19 × 11 × 11 cm3) plastic tanks until they were 7 months old (mean age in days±s.e.=204.10±1.48). As offspring within full-sibling families shared the same gestational and early rearing environment (up to 13 weeks), common environmental effects are likely to have contributed towards the dam variances, which were therefore not interpreted in this study (see below). The final dataset comprised 87 dam families from 29 sires, with variation within and among dam families in the number of male offspring assayed (mean±s.e. number of males per dam=5.74±0.21; range 1–19; total number of offspring assayed n=451). Eleven sire families were not included in the analysis owing to failed inseminations or insufficient numbers of dams producing offspring from each sire.
Sperm assays
Each male offspring was anaesthetized, carefully dried and placed on a glass slide under a dissecting microscope. A micropipette was used to add 40 μl of an extender medium (207 mM NaCl, 5.4 mM KCl, 1.3 mM CaCl2, 0.49 mM MgCl2, 0.41 mM MgSO4, and 10 mM Tris with pH 7.5) to the base of the male's gonopodium. The use of this extender ensured that spermatozeugmata (unencapsulated sperm bundles) remained intact and quiescent until they were used for the sperm assays (see Gardiner, 1978). Gentle pressure was then applied to each male's abdomen to expel strippable sperm into the extender medium (see Matthews et al., 1997). From this total sperm pool, eight spermatozeugmata were extracted to assess sperm viability using a live/dead sperm viability assay (Invitrogen, Molecular Probes, Eugene, OR, USA). The proportion of live sperm in each sample was then estimated from 200 sperm cells per sample (see Evans, 2009 for detailed methods of this assay).
Spermatozeugmata from the reserved sperm sample were activated with 40 μl of 150 mM KCl solution (see Billard and Cosson, 1990) containing 2 mg l−1 bovine serum albumin to prevent sperm from sticking to the glass slide (Pitcher et al., 2007). From these activated samples, three spermatozeugmata (comprising ca. 80 × 103 sperm cells) were immediately placed in a separate well of a 12-cell multitest slide (MP Biomedicals, Aurora, OH, USA) coated with a 1% polyvinyl alcohol (Wilson-Leedy and Ingermann, 2007). Each sample was then analysed using the CEROS Sperm Tracking program (Hamilton Thorne Research, Beverly, MA, USA). This software generated several estimates of sperm swimming velocity, including average path velocity (VAP; the average velocity of sperm cells over a smoothed cell path), straight line velocity (VSL; the average velocity on a straight line between the start and end point of the track) and curvilinear velocity (VCL; the actual velocity along the trajectory). As VAP was strongly positively correlated with both VSL (r=0.972) and VCL (r=0.952), I used just VAP in all subsequent analyses (see also Evans, 2009). (Note that results remained unchanged irrespective of the measure used.) The threshold value for defining static cells for VAP estimates was predetermined at 24.9 μm s−1. The CASA assays also generated measures of the amplitude of lateral head displacement, which is the mean width of the sperm's head oscillation, and beat cross frequency, which measures the frequency with which the sperm's head crossed the average path in either direction. CASA assays were based on a mean of 69.9±1.82 s.e. sperm tracked per sample.
Following the sperm velocity assays, 10 μl of each male's stripped ejaculate was reserved for the sperm length estimates. High resolution digital images of these samples were captured under × 400 magnification using a Leica DFC320 digital camera fitted to a Leica DM1000 microscope (Leica Microsystems Pty, North Ryde, Australia). Where possible, 15 spermatozoa were analysed per sample (mean number of sperm cells analysed per sample±s.e.=13.33±0.13; range 3–15). From these images, the length of the sperm head, midpiece, flagellum and total sperm length (=head+midpiece+flagellum) were estimated to be within 0.1 μm for each sperm cell using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Quantitative genetic analyses
All genetic analyses and associated s.e. estimates were conducted using linear mixed-effects models in ASReml 3.0 (VSN International Ltd, Hemel Hempstead, UK) (Gilmour et al., 2009). As the final dataset was unbalanced (that is there were variable numbers of male offspring tested among dam families, and variation in the number of dams per sire), variance components were estimated through restricted-maximum likelihood, which unlike least-squares methods does not place any demands on the balance of the data (Lynch and Walsh, 1998). For the estimation of genetic variances, univariate models were constructed with sire and dam identities included as random effects (dam IDs were nested within sire). The significance levels for sire variance components were estimated using likelihood-ratio tests, where full models (including the random effect of sires) were compared with reduced models (with sire terms removed). In each case the resulting G statistic, equivalent to -2 times the difference in log-likelihoods, between full and reduced models, was tested against a χ2 distribution (with d.f.=1). The variance components from the linear mixed-effects models were used to calculate additive genetic variances (VA) required for the estimation of narrow-sense heritabilities (Falconer and Mackay, 1996) and coefficients of additive genetic variation (Houle, 1992; see below). For heritabilities, separate estimates due to sires and dams were estimated, and these values were compared using the jackknife procedure suggested by Roff (2008). In this procedure, sire and dam heritability pseudovalues (generated by eliminating each sire family in successive heritability estimates) were compared using a one-sample t-test (Roff, 2008). The test has the useful property of testing for maternal and/or dominance effects, which will inflate dam heritabilities above values expected under purely additive genetic effects (Roff, 2008). Conversely, Y-linked genetic variation can be inferred where sire heritabilities significantly exceed those owing to dams, as sire heritabilities were calculated on the assumption of autosomal inheritance, which is twice the value expected if traits are completely Y-linked (Falconer and Mackay, 1996). Coefficients of phenotypic, additive genetic and residual variation were calculated using mean-standardized variance components, as described in the legend to Table 1 and outlined by Houle (1992).
Calculations of genetic correlations and their s.e. were also performed in ASReml using bivariate models with unconstrained variance matrices. Only the correlations due to covariance among sires were calculated, as dam estimates (derived from full-sibling families) were likely to be inflated by dominance variance, maternal and other environmental effects (Falconer and Mackay, 1996). To test the significance of each genetic correlation, the log-likelihood of a model with an unfixed covariance structure was compared with one in which the covariance was fixed at zero. The resultant (G) statistics were then tested against a χ2 distribution (with d.f.=1) using likelihood ratio tests (as above).
Results
Descriptive statistics
All ejaculate traits exhibited highly significant levels of additive genetic variation attributable to sires (Table 1). Narrow-sense heritabilities owing to sires exceeded those attributable to dams in all cases, and were generally high for all traits with the exception of amplitude of lateral head displacement and beat cross frequency, which exhibited moderate levels of heritability (Table 1; c. h2 values for sperm traits reviewed by Simmons and Moore, 2009). The very high heritabilities for all other traits is consistent with patterns of Y-linkage, especially where sire narrow-sense heritabilities exceeded dam values (note that heritabilities for sperm head length, total sperm length, VAP and sperm viability significantly exceeded dam values; see Table 1 for P-values associated with pairwise comparisons). In these cases, heritabilities are likely to be inflated by up to twice their actual value (see Materials and methods), which may account for why h2 values for sperm head, total sperm length and viability exceeded the theoretical maximum value of 1.0. Finally, coefficients of phenotypic and additive genetic variation were generally high for midpiece length, VAP, amplitude of lateral head displacement and sperm viability.
Patterns of genetic and phenotypic covariance
The patterns of genetic and phenotypic variation are summarized in Table 2. Contrary to expectation, midpiece length was strongly negatively genetically correlated with VAP. Furthermore, there were highly significant negative genetic correlations between both sperm head length and flagellum length and sperm viability. These patterns of genetic covariation between individual sperm components, and in vitro sperm performance are therefore likely to account for the negative correlations between total sperm length (which incorporates the head, midpiece and flagellum), and both VAP and viability (although note that only the latter genetic correlation was statistically significant; see bold values indicating significance in Table 2). Midpiece length was also positively genetically correlated with beat cross frequency, whereas VAP was positively genetically correlated with amplitude of lateral head displacement. Patterns of phenotypic covariation were largely consistent with these patterns of genetic covariation (see estimates below the diagonal in Table 2).
Discussion
This study reveals high levels of additive genetic and phenotypic variation underlying the expression of individual ejaculate characteristics in guppies. These patterns of variation are consistent with an emerging (but still small) body of literature revealing high levels of phenotypic, and genetic variation in ejaculate and fertility traits (quantitative genetic studies reviewed by Simmons and Moore, 2009).
The results from this study may shed some light on the possible location of genes that underlie ejaculate traits in guppies. The very high heritability estimates for all sperm length measures, as well as VAP and sperm viability, are consistent with the idea that much of the quantitative genetic variation underlying the expression of these traits is linked to the Y-chromosome. These patterns have parallels in the insect literature. For example, in the dung beetle, Onthophagus taurus, Simmons and Kotiaho (2002) also reported heritability estimates for sperm length that exceeded the theoretical maximum of 1.00. Furthermore, Joly et al. (1997) used hybrid crosses between Drosophila simulans and Drosophila sechellia to test the effects of the Y chromosome on sperm length. Specifically, they introgressed the Y chromosome of D. sechellia into the background of D. simulans, and found significant differences in sperm length between these introgressed strains and the pure D. simulans strain, indicating that Y-linked genes underlie variation in sperm length (Joly et al., 1997). Although in the present study, the patterns of genetic variation are suggestive of Y-linkage, other factors, such as the history of inbreeding in the focal population, may also influence patterns of genetic variation and consequently h2 (see, for example, review by van Buskirk and Willi, 2006). Importantly, the half-sibling breeding design is not sufficient for partitioning the Y-linked component from other sources of variance (X-linkage and additive genetic effects). For these purposes, genetic mapping studies are needed to link genes for ejaculate traits to specific chromosomes (for example, Johns and Wilkinson, 2007). Genomic resources have been developed in guppies (for example, Tripathi et al., 2009), making this species a good candidate for such approaches in the future.
Contrary to expectation, the present study revealed a highly significant negative genetic correlation between the length of the sperm's midpiece and VAP, and a weak negative phenotypic correlation between these traits. These patterns contrast with previous work on guppies revealing a positive phenotypic correlation between midpiece size and sperm velocity (Pitcher et al., 2007; Skinner and Watt, 2007). In these previous studies, however, the measure of midpiece ‘size’ differed from the measure employed in the current study. For example, in Pitcher et al.'s (2007) study, the midpiece was indistinguishable from the sperm's head, and thus, their measure was actually a composite measure of head and midpiece length. Nevertheless, when I incorporated the equivalent measure (sperm head+midpiece) in my analysis, the negative genetic correlation between this composite measure and VAP was even stronger than that for the midpiece alone (rG=−0.91±0.15 s.e.; cf. Table 2). In Skinner and Watt's (2007) study, the authors reported midpiece area (rather than length) and sperm swimming velocity was evaluated 15 min after sperm activation. Thus, equivalent measures are unavailable in the present study for comparison. Nevertheless, the present study and Pitcher et al.'s (2007) analysis are comparable, and the differences between the two studies may either reflect different methodologies for estimating sperm velocity (for example, CASA was not used in Pitcher et al.'s study and the number of sperm tracked per male was lower than those measured here, varying from 12–26 across their samples) or biologically meaningful differences between the two populations (for example, see Pitcher and Evans, 2001). An important next step is to determine how midpiece size (and other measures of sperm length) impact on male fertility and competitive fertilization success to determine the extent to which the patterns uncovered in both studies may influence the evolution of these traits.
Although the negative correlations between midpiece length and VAP appear to be at odds with previous work on guppies, these findings are consistent with work on other taxa. For example, Malo et al. (2006) reported a negative phenotypic correlation between midpiece length and VAP in red deer, whereas Mossman et al. (2009) have recently reported a negative genetic correlation between midpiece length and sperm velocity in zebra finches. Similarly, Firman and Simmons (2008) showed that in house mice sperm length (including the midpiece) was negatively associated with competitive fertilization success, although in that species midpiece length appears to be positively associated with sperm swimming velocity (Firman and Simmons, 2010). Interestingly, in house mice sperm velocity was negatively associated with sperm longevity (Firman and Simmons, 2010), suggesting that selection favours an optimum length that maximizes sperm velocity and longevity. In red deer, sperm swimming velocity was an important predictor of (non-competitive) fertilization rates (Malo et al., 2005), suggesting a potential fitness benefit of producing relatively short midpieces. However, as with guppies, the role of the midpiece in mediating competitive fertilization success has yet to be established in any of these species.
Interestingly, the present study revealed no evidence for either phenotypic or genetic integration between the length of the sperm midpiece and other measures of sperm length, suggesting that the relationship between this trait and swimming velocity is independent of any other sperm length measures. Indeed, midpiece length was not significantly genetically correlated with flagellum length, although the sign of both phenotypic and genetic correlations is consistent with previous work, revealing a negative correlations between midpiece length and flagellum length, both in guppies (phenotypic correlation; Skinner and Watt, 2007) and zebra finches (genetic correlation; Birkhead et al., 2005). In the zebra finch example, however, flagellum length is more closely associated with total sperm length than midpiece size, and therefore the strong positive genetic correlation between flagellum length and sperm swimming velocity will likely favour longer, not shorter, sperm in this species (Mossman et al., 2009). By contrast, the present study suggests that all components of sperm length are negatively genetically associated with at least one aspect of in vitro sperm performance (VAP or viability), suggesting that short sperm will be favoured in sperm competition (see also Firman and Simmons, 2008; Gage and Morrow, 2003; García-González and Simmons, 2007).
The likely benefit of producing shorter sperm in guppies is further underscored by the negative genetic correlations between sperm viability, and the length of both the sperm's head and flagellum. These patterns of negative covariance were striking for both traits, and suggest that these two components of sperm length (which are easily distinguishable from the midpiece in guppies) are strongly genetically integrated with the viability of sperm. These patterns are apparently without precedent in the quantitative genetic literature, although Dowling et al. (2007) reported a near-significant negative correlation between sperm viability and sperm length across nuclear lineages of the seed beetle Callosobruchus maculatus. However, as with guppies, the evolutionary significance of the relationship between sperm length and sperm viability has yet to be determined in Callosobruchus maculatus.
Finally, this study serves as a prospective analysis of quantitative genetic variation in ejaculate traits, which is required for addressing hypotheses proposing that females benefit indirectly from polyandry by producing sons that inherit genes that improve their prospects during sperm competition (Curtsinger, 1991; Yasui, 1997). These so-called ‘good-sperm’ and ‘sexy-sperm’ models predict that by mating with multiple males, the ensuing contest among rival male ejaculates ensures that fertilization will favour individuals that transmit genes for improved sperm performance to their male offspring (Sivinski, 1984; Harvey and May, 1989; Curtsinger, 1991; Keller and Reeve, 1995; Yasui, 1997). Both models therefore, share the prediction that there must be sufficient additive genetic variance attributable to sires underlying the expression of functionally important ejaculate traits (Evans and Simmons, 2008). In this respect, the results from the current study add to a growing number of studies offering explicit support for this component of ‘good-sperm’ and ‘sexy-sperm’ theory (reviewed in Evans and Simmons, 2008; Simmons and Moore, 2009), but also highlight potentially important genetic constraints that may impede the evolutionary trajectories of individual sperm traits (see also Birkhead et al., 2005; Moore et al., 2004).
In summary, the results from this paper underscore the complex genetic relationships that underlie ejaculate traits reported elsewhere (Moore et al., 2004; Birkhead et al., 2005). They also add to the emerging picture, that ejaculates are characterized by extremely high levels of phenotypic and genetic variation, thus making them potential targets of post-mating sexual selection. Nevertheless, understanding the functional significance of these patterns remain important challenges for the future. As Pizzari and Parker (2009) have recently suggested, the tools of multivariate selection analyses (see Lande and Arnold, 1983) are likely to prove helpful for exploring such patterns by unveiling the direction, strength and form of selection acting on these multifarious traits.
References
Billard R, Cosson MP (1990). The energetics of fish sperm motility. In: Gagnon C (ed.). Controls of Sperm Motility: Biological and Clinical Aspects. CRC Press, Inc: Boca Raton. pp 153–173.
Birkhead TR, Martínez JG, Burke T, Froman DP (1999). Sperm mobility determines the outcome of sperm competition in the domestic fowl. Proc R Soc Lond B 266: 1759–1764.
Birkhead TR, Møller AP (1998). Sperm Competition and Sexual Selection. Academic Press: San Diego.
Birkhead TR, Pellatt EJ, Brekke P, Yeates R, Castillo-Juarez H (2005). Genetic effects on sperm design in the zebra finch. Nature 434: 383–387.
Curtsinger JW (1991). Sperm competition and the evolution of multiple mating. Am Nat 138: 93–102.
Denk AG, Holzmann A, Peters A, Vermeirssen ELM, Kempenaers B (2005). Paternity in mallards: effects of sperm quality and female sperm selection for inbreeding avoidance. Behav Ecol 16: 825–833.
Dowling DK, Nowostawski AL, Arnqvist G (2007). Effects of cytoplasmic genes on sperm viability and sperm morphology in a seed beetle: implications for sperm competition theory? J Evol Biol 20: 358–368.
Elgee KE, Evans JP, Ramnarine IW, Pitcher TE (2010). Geographic variation in sperm traits reflects predation risk and natural rates of multiple paternity in the guppy. J Evol Biol 23: 1331–1338.
Evans JP (2009). No evidence for sperm priming responses under varying sperm competition risk or intensity in guppies. Naturwissenschaften 96: 771–779.
Evans JP (2010). Quantitative genetic evidence that males trade attractiveness for ejaculate quality in guppies. Proc R Soc Lond B 277: 3195–3201.
Evans JP, Simmons LW (2008). The genetic basis of traits regulating sperm competition and polyandry: can selection favour the evolution of good- and sexy-sperm? Genetica 134: 5–19.
Falconer DS, Mackay TFC (1996). Introduction to Quantitative Genetics. Longman Group Ltd: London.
Firman RC, Simmons LW (2008). Polyandry, sperm competition, and reproductive success in mice. Behav Ecol 19: 695–702.
Firman RC, Simmons LW (2010). Sperm midpiece length predicts sperm swimming velocity in house mice. Biol Lett 6: 513–516.
Fitzpatrick JL, Garcia-Gonzalez F, Evans JP (2010). Linking sperm length and velocity: the importance of intramale variation. Biol Lett (e-pub ahead of print; doi:10.1098/rsbl.2010.0231).
Fitzpatrick JL, Montgomerie R, Desjardins JK, Stiver KA, Kolm N, Balshine S (2009). Female promiscuity promotes the evolution of faster sperm in cichlid fishes. Proc Natl Acad Sci USA 106: 1128–1132.
Gage MJG, Macfarlane CP, Yeates S, Ward RG, Searle JB, Parker GA (2004). Spermatozoal traits and sperm competition in Atlantic salmon: relative velocity is the primary determinant of fertilization success. Curr Biol 14: 44–47.
Gage MJG, Morrow EH (2003). Experimental evidence for the evolution of numerous, tiny sperm via sperm competition. Curr Biol 13: 754–757.
García-González F, Simmons LW (2005). Sperm viability matters in insect sperm competition. Curr Biol 15: 271–275.
García-González F, Simmons LW (2007). Shorter sperm confer higher competitive fertilization success. Evolution 61: 816–824.
Gardiner DM (1978). Utilization of extracellular glucose by spermatozoa of two viviparous fishes. Comp Biochem Physiol 59A: 165–168.
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009). ASReml User Guide Release 30. VSN International Ltd: Hemel Hempstead.
Gomendio M, Roldan RS (2008). Implications of diversity in sperm size and function for sperm competition and fertility. Int J Dev Biol 52: 439–447.
Harvey PH, May RM (1989). Out for the sperm count. Nature 337: 508–509.
Houle D (1992). Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204.
Humphries S, Evans JP, Simmons LW (2008). Sperm competition: linking form to function. BMC Evol Biol 8: 319.
Hunter FM, Birkhead TR (2002). Sperm viability and sperm competition in insects. Curr Biol 12: 121–123.
Johns PM, Wilkinson GS (2007). X chromosome influences sperm length in the stalk-eyed fly Cyrtodiopsis dalmanni. Heredity 99: 56–61.
Joly D, Bazin C, Zeng LW, Singh RS (1997). Genetic basis of sperm and testis length differences and epistatic effect on hybrid inviability and sperm motility between Drosophila simulans and D sechellia. Heredity 78: 354–362.
Keller L, Reeve HK (1995). Why do females mate with multiple males? The sexually selected sperm hypothesis. Adv Stud Behav 24: 291–315.
Kotiaho JS, Simmons LW, Hunt J, Tomkins JL (2003). Males influence maternal effects that promote sexual selection: a quantitative genetic experiment with dung beetles Onthophagus taurus. Am Nat 161: 852–859.
Lande R, Arnold SJ (1983). The measurement of selection on correlated characters. Evolution 37: 1210–1226.
Liljedal S, Rudolfsen G, Folstad I (2008). Factors predicting male fertilization success in an external fertilizer. Behav Ecol Sociobiol 62: 1805–1811.
Lindholm AK, Breden F, Alexander HJ, Chan WK, Thakurta SG, Brooks R (2005). Invasion success and genetic diversity of introduced populations of guppies Poecilia reticulata in Australia. Mol Ecol 14: 3671–3682.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc: Sunderland.
Magurran AE (2005). Evolutionary Ecology: The Trinidadian Guppy. Oxford University Press: Oxford.
Malo AF, Garde JJ, Soler AJ, Garcia AJ, Gomendio M, Roldan ERS (2005). Male fertility in natural populations of red deer is determined by sperm velocity and the proportion of normal spermatozoa. Biol Reprod 72: 822–829.
Malo AF, Gomendio M, Garde J, Lang-Lenton B, Soler AJ, Roldan ERS (2006). Sperm design and sperm function. Biol Lett 2: 246–249.
Matthews IM, Evans JP, Magurran AE (1997). Male display rate reveals ejaculate characteristics in the Trinidadian guppy Poecilia reticulata. Proc R Soc Lond B 264: 695–700.
Moore PJ, Harris WE, Montrose VT, Levin D, Moore AJ (2004). Constraints on evolution and postcopulatory sexual selection: trade-offs among ejaculate characteristics. Evolution 58: 1773–1780.
Mossman J, Slate J, Humphries S, Birkhead TR (2009). Sperm morphology and velocity are genetically codetermined in the zebra finch. Evolution 63: 2730–2737.
Parker GA (1970). Sperm competition and its evolutionary consequences in the insects. Biol Rev 45: 525–567.
Pilastro A, Simonato M, Bisazza A, Evans JP (2004). Cryptic female preferences for colorful males in guppies. Evolution 58: 665–669.
Pitcher TE, Evans JP (2001). Male phenotype and sperm number in the guppy (Poecilia reticulata). Can J Zool 79: 1891–1896.
Pitcher TE, Rodd FH, Rowe L (2007). Sexual colouration and sperm traits in guppies. J Fish Biol 70: 165–177.
Pizzari T, Parker GA (2009). Sperm competition and sperm phenotype. In: Birkhead TR, Hosken DJ, Pitnick S (eds). Sperm Biology: An Evolutionary Perspective. Academic Press: Burlington, MA, pp 207–245.
Roff DA (2008). Comparing sire and dam estimates of heritability: jackknife and likelihood approaches. Heredity 100: 32–38.
Simmons LW, Kotiaho JS (2002). Evolution of ejaculates: patterns of phenotypic and genotypic variation and condition dependence in sperm competition traits. Evolution 56: 1622–1631.
Simmons LW, Moore AJ (2009). Evolutionary quantitative genetics of sperm. In: Birkhead TR, Hosken DJ, Pitnick S (eds). Sperm Evolution: An Evolutionary Perspective. Academic Press: Burlington, MA, pp 405–434.
Sivinski J (1984). Sperm in competition. In: Smith RL (ed). Sperm Competition and the Evolution of Animal Mating Systems. Academic Press: Orlando, FL.
Skinner AMJ, Watt PJ (2007). Phenotypic correlates of spermatozoon quality in the guppy, Poecilia reticulata. Behav Ecol 18: 47–52.
Skjaeraasen JE, Mayer I, Meager JJ, Rudolfsen G, Karlsen O, Haugland T et al. (2009). Sperm characteristics and competitive ability in farmed and wild cod. Mar Ecol Prog Ser 375: 219–228.
Snook RR (2005). Sperm in competition: not playing by the numbers. Trends Ecol Evol 20: 46–53.
Snook RR, Bacigalupe LD, Moore AJ (2010). The quantitative genetics and coevolution of male and female reproductive traits. Evolution 64: 1926–1934.
Tourmente M, Gomendio M, Roldan ERS, Giojalas LC, Chiaraviglio M (2009). Sperm competition and reproductive mode influence sperm dimensions and structure among snakes. Evolution 63: 2513–2524.
Tripathi N, Hoffmann M, Willing EM, Lanz C, Weigel D, Dreyer C (2009). Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc R Soc Lond B 276: 2195–2208.
van Buskirk J, Willi Y (2006). The change in quantitative genetic variation with inbreeding. Evolution 60: 2428–2434.
Wilson-Leedy JG, Ingermann RL (2007). Development of a novel CASA system based on open source software for characterization of zebrafish sperm motility parameters. Theriogenology 67: 661–672.
Yasui Y (1997). A ‘good-sperm’ model can explain the evolution of costly multiple mating by females. Am Nat 149: 573–584.
Acknowledgements
I am especially grateful to Cameron Duggin for help with animal rearing, general maintenance and the sperm viability assays, and John Trainer for taking sperm length measurements. I also thank Paco García-González, Leigh Simmons and three anonymous reviewers for comments on a previous draft of the manuscript, Andrea Pilastro for helpful advice with ASReml, and the Australian Research Council for financial support. This research was performed according to guidelines governing the use of animals in research and was approved under the University of Western Australia's animal ethics license 05/100/513.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Evans, J. Patterns of genetic variation and covariation in ejaculate traits reveal potential evolutionary constraints in guppies. Heredity 106, 869–875 (2011). https://doi.org/10.1038/hdy.2010.130
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.130
Keywords
This article is cited by
-
Maternal-by-environment but not genotype-by-environment interactions in a fish without parental care
Heredity (2018)
-
Rapid evolution of sperm length in response to increased temperature in an ectothermic fish
Evolutionary Ecology (2014)
-
Functional variation of sperm morphology in sticklebacks
Behavioral Ecology and Sociobiology (2014)