Abstract
Grouping provides many potential benefits to individuals in terms of foraging and anti-predator protection. However, it has been suggested that individuals could gain additional benefits in terms of indirect fitness by grouping with kin. Surprisingly, the genetic composition of wild fish shoals and the importance of kin-associated shoaling remain poorly understood. The Trinidadian guppy (Poecilia reticulata) has life history traits that might promote kin structure of shoals such as internal fertilisation and small brood size in contrast to many other fish species. Even though previous studies did not find any indication of kin structure in shoals of adult guppies, it is possible that related juveniles remain together in shoals, partly because of lower mobility and because the advantages of kin association may change with age. Using 10 microsatellite markers, we conducted a genetic analysis on 40 shoals from four populations. Pair-wise relatedness was inferred using a modified version of the software package COLONY and permutation tests were conducted to test the hypothesis that kin occur together in juvenile shoals more often than expected by chance. The frequency of sib dyads among juveniles within shoals was significantly larger than that between shoals in two high predation populations but not in two low predation populations. This finding contributes to the understanding of factors underlying shoal composition and highlights the potential of recent methodological advances for detecting such relationships.
Similar content being viewed by others
Introduction
Group living is ubiquitous across the animal kingdom and the associated fitness benefits have been well explored (Krause and Ruxton, 2002). These benefits include anti-predation protection and increased foraging efficiency and these behaviours are assumed to play a major role in the social organisation of animals (Pitcher and Parrish, 1993). Associating with relatives may increase an individual's fitness because kin are likely to be more cooperative when engaging in risky behaviours such as predator inspection (Milinski, 1987; Ward and Hart, 2003), providing indirect fitness benefits to individuals (Hamilton, 1964). Previous studies found that individuals are choosy regarding shoal mates during the approach towards a predator and indicated that fish can gain important information during predator inspection behaviour regarding the nutritional state of predators (Dugatkin, 1997; Croft et al., 2006). These assortative associations are thought to be an important prerequisite for the evolution of cooperative behaviour (Croft et al., 2009).
Genetic relatedness among individuals in social-groups is an important factor in understanding kin selection and the evolution of altruism (Hamilton, 1964; Foster et al., 2006). Association in kin-groups has been well described across a range of taxa from primitive Eukaryotes, both unicellular and multicellular forms (Gilbert et al., 2007), to birds and mammals (Emlen and Wrege, 1988; Packer et al., 1991). However, there are few studies of the role of kin association in fish and the relative importance of kin-associated shoaling remains equivocal (Krause et al., 2000; Griffiths and Ward, 2006).
There is evidence of kin recognition in a number of fish species from studies conducted in laboratory conditions (Arnold, 2000; Hiscock and Brown, 2000; Frommen and Bakker, 2004; Frommen et al., 2007; Hain and Neff, 2007). Many investigations of kin recognition in fish have focused on juvenile salmonids and have shown that salmonids are able to recognise kin (see Olsén, 1999 for a review). Experiments using released PIT (Passive Integrated Transponders) tagged smolts of laboratory reared salmon, have shown that siblings swim spatially closer together than unrelated fish during their seaward migration, supporting the hypothesis that smolts migrate in kin-structured groups (Olsen et al., 2004). These studies provide reliable evidence for kin recognition and kin preference both under laboratory and semi-field conditions. However, more generally, difficulties exist in detecting kinship in fish shoals in the wild (see Krause et al., 2000 for a review, Gerlach et al., 2001; Russell et al., 2004; Fraser et al., 2005) because there are a number of methodological issues which complicate the conclusions that can be drawn from them. Early studies (Avise and Shapiro, 1986; Naish et al., 1993; Peuhkuri and Seppa, 1998) may not have used sufficiently informative genetic markers to demonstrate that more kin were found within shoals than expected by chance. Since numerous, highly polymorphic microsatellite loci are now available for many species, the power of markers need not be a limitation in current relatedness studies (Luikart and England, 1999; Blouin, 2003). However, an additional issue in recent investigations using microsatellites is that kin-associated shoaling may have been confounded with spatial genetic structure within a population (Pouyaud et al., 1999). Another problem of studying free-ranging shoals is that it is necessary to capture an entire shoal to be able to draw firm statistically testable conclusions regarding its size and composition (Krause et al., 2000). This has been an issue in several recent studies that either find no kin association (Herbinger et al., 1997), or do suggest that kin association occurs (Gerlach et al., 2001; Fraser et al., 2005).
In this study, we used guppies to investigate kin assortment in fish shoals. Guppies (Poecilia reticulata) have become important model organisms in evolutionary biology and behavioural ecology, especially regarding research topics such as sexual selection and group living (Houde, 1997; Magurran, 2005; Breden, 2006). They have internal fertilisation, give birth to living young, and juveniles from the same brood demonstrate a high tendency to remain together in the laboratory (Magurran and Seghers, 1990). Adult guppies are known to have a preference to shoal with familiar individuals and this preference develops after a period of 12 days (Griffiths and Magurran, 1997, 1999). However, the potential to confound familiarity with kinship preferences can be reduced when working with fries or juveniles because they have had less time to form familiarity preferences. There is evidence that juvenile guppies prefer to shoal with kin rather than non-kin and with full sibs rather than half sibs in the laboratory (Hain and Neff, 2007; Evans and Kelley, 2008) and here we test whether this is also true in the field. Russell et al. (2004), however, found no significant kin structure in wild adult Trinidadian guppy shoals from two high predation rivers. This study used RELATEDNESS (implementing the algorithm described by Queller and Goodnight (1989)) to calculate the relatedness coefficient (r) between pairs of fish. Because all estimators of r suffer from large sampling errors (Lynch and Ritland, 1999 see below) except when an extremely large number of polymorphic markers are used, it is possible that biologically significant relatedness within shoals might be present but not detected by Russell et al. (2004) or similar studies.
When the number of possible candidate relationships (such as unrelated, full-sibs and half-sibs with r=0, 0.5 and 0.25, respectively) is small and these candidate relationships are well differentiated in the extent and /or pattern of relatedness, relationship inference is generally much more powerful than relatedness estimation. This is because r can take any value in a continuous scale from 0 to 1 in relatedness estimation, but can take just a few well separated values in relationship inference. Furthermore, while relatedness estimation uses the genotype data from only two individuals, relationship inference can use genotype information from multiple relatives jointly in a likelihood framework (Wang, 2004). As a result, the contrast in accuracy between relatedness and relationship inference becomes more evident with an increasing family size. Therefore, rather than estimating and comparing relatedness within and between shoals, it is preferable to infer relationships among all individuals within the sample, using maximum likelihood methods (Wang, 2004), and then test whether closely related individuals, such as full or half-sibs, are more frequent within than between shoals. Here, we use this approach to test the hypothesis that closely related individuals tend to be found together in juvenile guppy shoals in the wild.
Materials and methods
Shoal sampling
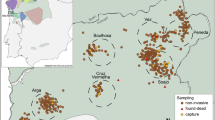
Entire shoals of juvenile guppies were collected using seine nets (the mesh size was 1 × 1 mm2 ) from four populations (upper Aripo, lower Aripo, Arouca and Paria) in May and June 2007 (Table 1). We observed fish from each shoal before capturing them to ensure they were sampled as a complete shoal. All fish of total body length (TL) between 7 and 17 mm were categorised as juveniles and were estimated to be less than 50 days old (Chapman et al., 2008; Ledesma and McRobert, 2008). For each population, five transects were sampled along the river and the position along the transect where each shoal was caught was recorded. Each transect was 10 m long and, where possible, the transects were adjacent and in the same direction, with no gap between transects. However, due to the presence of small waterfalls and fast currents in some rivers, gaps between transects did exist. Overall, the distance from the start of the first transect to the end of the last ranged from 50 to 100 m. All collected fish were euthanized by an overdose of methanesulfonate (MS-222) and then were preserved in 80% ethanol at ambient temperature for subsequent genetic analysis.
Genetic analysis
Genomic DNA was extracted from the posterior part of each individual using the ammonium acetate precipitation method (Nicholls et al., 2000). Microsatellite multiplexing was used in this study because it can significantly reduce the time and expense of genotyping (Neff et al., 2000). Thirteen loci were chosen on the basis of possessing high heterozygosity and a low expected null allele frequency from the many loci described for guppies: one mixed di-tetranucleotide locus, Pr92, (Becher et al., 2002), one di- and three tetranucleotide loci, AG4, AATG2, AGAT10, and AGAT11, (Olendorf et al., 2004), four tetranucleotide loci, Pre8, Pre9, Pre15, and Pre26, (Paterson et al., 2005), two dinucleotide loci, Pret-27 and Pret-32, (Watanabe et al., 2003) and one dinucleotide locus and one trinucleotide locus from GenBank( AF170707 and AF164205, respectively). The Qiagen multiplex PCR method was used throughout this study (Engel et al., 2003). Final PCR amplification for all primers was performed using a DNA Engine Tetrad 2 thermal cycler (MJ Research) with the program: 95 °C for 15 min; 35 cycles of 94 °C for 30 s, specific primer annealing temperature (61 °C) for 90 s, 72 °C for 60 s and one final extension step of 60 °C for 30 min. Following optimisation for all four populations, three loci were dropped due to difficulties when scoring alleles consistently, leaving 10 polymorphic loci displaying 7–36 alleles (Table 2). These 10 loci were confirmed as unique within this set by comparing their sequences using NBLAST software (Altschul et al., 1997). Also, 10 adult males and females, descendants from the Tacarigua population, were tested for each locus to check for the presence of two alleles in both sexes. Genotypes of all individuals were assigned on an ABI 3730 DNA Analyzer using GENEMAPPER software v3.7 (Applied Biosystems). All individuals used for further analysis had at least 7 loci amplified and scored successfully (97.9 % for 10 loci, 1.5 % for 9 loci, 0.3 % for 8 loci and 0.3 % for 7 loci, respectively, see Supplementary information).
Statistical analysis
Data preparation and analysis were carried out using GENALEX (Peakall and Smouse, 2006), unless otherwise specified. Tests for departure from Hardy-Weinberg equilibrium (HWE; 100 batches and 1000 iterations per batch) for each locus in each population and pairwise tests for linkage disequilibrium (10 000 batches and 10 000 iterations per batch) were calculated in GENEPOP v3.4 (Raymond and Rousset, 1995) and corrected for multiple comparisons (Rice, 1989). Genetic variation was calculated at two hierarchical levels of population structure. Genetic variation among the four populations was estimated using an analysis of molecular variance (AMOVA) (Michalakis and Excoffier, 1996). Isolation by distance was tested among shoals within populations (Rousset, 1997) using a Mantel test (Mantel, 1967) (cumulative straight-line distance along the river between shoals in each population, implemented in R version 2.5.0; APE package (Paradis et al., 2004) with 1000 permutations). Since all FST values were below 0.1 (see below), it was not necessary to linearise the divergence measure, using FST/(1−FST), and, since the habitat is linear, distance was not log-transformed (Rousset, 1997).
Pairwise relatedness, r (QG) (Queller and Goodnight, 1989), within and between shoals, was calculated separately for each population using SPAGEDi version 1.2 (Hardy and Vekemans, 2002). Departure from the null hypothesis of equal relatedness within and between shoals was tested by permuting the relatedness matrix 1000 times using GENSTAT v10 (VSN International) for each population.
Relationships were estimated in two ways. We used ML-Relate (Kalinowski et al., 2006) to estimate the number of full and half-sib pairs within and between shoals in each population. The significance of departures from equal distribution of sib pairs was tested by permutation of the matrix as for relatedness. We also tested the sensitivity to individual microsatellite loci by leaving out one locus at a time and recalculating the relationship matrix. Finally, we inferred relationships using the program COLONY (Wang, 2004) extended to allow for both polyandry and polygyny (Wang and Santure, 2009). The program searches for the maximum likelihood partition of a sample of individuals into full- and half-sib clusters. It is more powerful than the pairwise approach because more information on entire families rather than just pairs of individuals is extracted and utilised (Wang, 2007; Wang and Santure, 2009). Bootstrapping (over individuals) was used to assess whether the average relatedness, calculated from the inferred sibship assignments, or the frequency of sibship was significantly greater within than between shoals, for each population. For this analysis, the typing error rate was set at 0.01 (Blouin, 2003).
Results
Genetic diversity
All loci for all four populations showed polymorphism (Table 2). Exact tests for linkage disequilibrium yielded no significant value for any pair of the loci (P>0.05). All loci had two alleles in both sexes. In the lower Aripo population, AGAT10, Pret-27, and Pre26 were suspected to have null alleles present due to departures from Hardy-Weinberg expectations, whereas Pret27, Pre8 and Pre15 were suspected to have null alleles in Arouca. However, no locus departed significantly from Hardy-Weinberg expectations in either the upper Aripo or Paria populations.
Genetic differentiation within and among populations
The AMOVA showed highly significant genetic variation among populations (FST=0.237, P<0.001). The pair-wise FST values between populations ranged from 0.082 to 0.385. Compared with the other three populations, the Paria sample is more differentiated than the rest. The AMOVA tests within each population detected low but significant genetic variation among shoals within three populations but not within the Upper Aripo: Paria (FST=0.014, P=0.024), Upper Aripo (FST=0.002, P=0.310), Lower Aripo (FST=0.009, P= 0.019) and Arouca (FST=0.014, P=0.002). These results were not qualitatively different when loci showing evidence for the presence of null alleles were removed from the calculations.
The Mantel tests revealed no significant isolation by distance no matter which population was used for the analysis (r=−0.240–0.274, P>0.05). These results did not change when loci with suspected null alleles were excluded.
Genetic relatedness and relationship within and between shoals
When using the r (QG) estimator to calculate relatedness, the mean was close to zero both within and between shoals for each population. Based on the permutation tests for each population, the null hypothesis of equal relatedness within and between shoals (Table 3) was not rejected.
When ML-Relate was used to estimate pairwise relationships, null alleles were specified for those loci listed above for each population before calculating the relationships. All first degree relatives identified by this program were categorised as full sib pairs since parent-offspring pairs could not occur amongst juveniles. Permutation tests were used to evaluate whether the frequency of sib pairs within shoals was greater than expected from random assortment of individuals among shoals, separating full sib and half sib categories. The expectations were exceeded significantly only in the two high-predation populations, lower Aripo and Arouca, and not in the two low predation populations (Table 4).
To see whether the observed excess of full or half sibs within shoals was sensitive to the loci included, we analysed data sets in ML-Relate leaving out one locus at a time. The proportions of full and half-sib relationships that occurred within shoals were calculated and compared with an expected value based on the overall proportion of within-shoal pairs. In the low-predation populations, the ranges of values included the expected values, or were lower (Paria: expected value=0.105, half-sibs=0.100–0.111 and full-sibs=0.096–0.155; upper Aripo: expected value=0.083, half-sibs=0.068–0.081 and full-sibs=0.082–0.110), whereas in the high predation populations this was not the case for full-sibs ( lower Aripo: expected value=0.089, half-sibs=0.080–0.099 and full-sibs=0.118–0.191; Arouca: expected value=0.117, half sibs=0.104–0.141 and full-sibs=0.240–0.302). Thus our conclusion that closely related pairs of individuals occur more frequently than expected within shoals is not sensitive to the effects of individual loci, at least in the case of full-sibs.
Using the relationships inferred by the COLONY program, we calculated the average relatedness (R) and frequency of sibship (P) within and between shoals, and employed bootstrapping to obtain confidence intervals on the difference in R and P within and between shoals under the null hypothesis. It was found that both P and R were higher within than between shoals for the lower Aripo and Arouca high-predation populations (observed differences fall outside the 95% confident intervals). There was no significant difference for the upper Aripo and Paria samples which are from low-predation populations (Table 5).
Discussion
We have found evidence to support the idea that there is kin structure in juvenile guppy shoals from high-predation populations but not low-predation ones. This pattern is unaffected by spatial genetic structure among shoals which was very low within our study sites.
Our AMOVA results (pairwise FST: Paria VS Arouca=0.281, Paria VS upper Aripo=0.385 and Paria VS lower Aripo=0.317) support the conclusion from a previous study on mtDNA that guppies from Paria, located in the Northern drainage, are more differentiated than the other three populations which are located in the Caroni drainage (Fajen and Breden, 1992). Guppies from the same tributary, upper Aripo and lower Aripo, exhibited little genetic differentiation (FST=0.082). This similarity may be due to high natural gene flow between these two populations but it could also have been influenced by transplantation of guppies from the lower Aripo to upper Aripo in 1976 (Endler, 1980). This introduction of guppies has affected not only gene flow, but also morphology, especially male colour pattern (Endler, 1980). We found evidence of low genetic differentiation among shoals within each of three populations but no evidence for isolation by distance. Differentiation is expected if there is kin assortment and the lack of spatial pattern suggests that this is a likely explanation for the weak structure observed.
Our study reveals that juvenile guppies in the wild tend to be found in shoals with related individuals in high-predation sites but not low-predation sites. This is the case even though the overall frequency of sibs in our samples was similar across high and low predation populations, both between and within shoals. This finding is likely to exclude the possibility that the observed pattern was due to predation on individuals that move between shoals. Our result is consistent with a previous study which found that female guppies from a high-predation population showed high site fidelity (Croft et al., 2003). This behaviour may cause them to give birth to offspring from the same brood in the same place. Also, our finding fits with an earlier study which detected no kin structure in adult guppies from a low-predation population (Hain and Neff, 2007). However, our results contradict Russell et al.'s (2004) observation of no kin structure among shoals in two populations from high-predation regimes. This discrepancy may be explained by the fact that we investigated juveniles (instead of adults) and/or it could be due to methodological differences. These methodological issues, combined with different methods of data analysis, could mean that the level of kin assortment that we observed in juvenile guppies has been missed in previous studies and may also exist in adult guppies in high-predation rivers. This topic should be revisited in future studies and more sites need to be analysed in order to test the relationship between predation level and kin assortment more thoroughly. Also, it might be more powerful to compare pairs of populations from the same tributary like we did with the lower Aripo and upper Aripo. Our finding from these two populations implies that both geographical variation and predation level may influence the expression of kinship assortment in guppies.
Low predation populations of Trinidadian guppies tend to have smaller brood sizes than high predation populations (reviewed in Magurran, 2005). Juveniles from both populations may tend to stay together with their brood after birth, perhaps until they become mature, but some broods will fuse to form larger shoals and shoals are likely to divide and exchange individuals, diluting the initial pattern of kin-relationships. If similar numbers of individuals per brood from both low-predation and high-predation populations leave the shoal but remain in the population, they would have a greater effect on our analysis for the low predation population than high predation population, because they represent a higher proportion of the sibship. However, we have shown that there were larger numbers of siblings among the shoals for low predation populations than for high-predation populations (Table 6), contrary to expectation. Indeed, total sibships were larger for low predation populations in our data but this could be because we sampled a higher proportion of the total population in these sites. Another possibility is that juvenile guppies from high predation populations may not survive to find another shoal because a previous study found that they suffered higher mortality rates than adults in these populations (Reznick et al., 1996).
Comparing the different analytic methods to quantify kinship in guppies, our empirical study confirmed that the maximum likelihood approach to relationship inference has greater power to detect kinship than the relatedness approach. Relatedness quantifies the extent that two individuals are genetically related due to the sharing of common ancestors. In an outbred population, its legitimate value lies in the range between 0 (unrelated dyads) and 1 (monozygotic twins), but its estimated value frequently lies well outside of this range except when it is forced into the range by truncation (as in the likelihood methods) or when extremely informative markers are utilised. No matter which relatedness estimator is used, therefore, the estimates are quite noisy and the estimation errors are expected to be high with just 10 microsatellites (Blouin, 2003). Any difference in average relatedness within and between shoals is thus easily blurred by the high sampling variance in relatedness estimates. By comparison, relationships are more reliably inferred, especially when the number of candidate relationships is small and these candidates are well differentiated (Blouin, 2003). In an attempt to understand the nature of the kin-groups, we counted the number of sets of individuals connected by at least a half-sib relationship (as inferred by ML-Relate) and the number of individuals in each set, within each shoal. The lower Aripo and Arouca populations contained an average number of groups of relatives (±s.e.) of 1.73±0.33 and 1.44±0.18, respectively, each one of which contained an average of 3.56±0.36 and 7.17±1.54 fish and the average percentages of fish in related groups were 40±6 and 62±7%.
The COLONY software operates differently (Wang, 2004; Wang and Santure, 2009): by attempting to reconstruct the pedigree of sampled individuals, it defines groups of sibs sharing either one or both parents. This analysis gives a superficially different view of the pattern of relationships (Table 6) suggesting that the average size of groups of sibs in our samples is around 4-8 fish in total, with 10–20% of sibs occurring within shoals. However, the inferred groups of half-sibs can be interrelated. For example, individual B may be a paternal half-sib of individual A and maternal half-sib of individual C. Although A and C are unrelated genetically because they do not share any parent, they are connected through the parents of B. If overlapping sets of half-sibs, connected by different parents, are considered, networks of related individuals similar in size to those detected by ML-Relate can be detected. Maternal half-sibships are more likely to be important than paternal half-sibships, if the kinship structure is driven by brood-members tending to stay together, but we are not able to separate these relationships with our data.
Free-ranging animal groups are frequently assorted by a range of phenotypic characters. For example, it is well known that individuals may make active decisions to join groups based on a number of characteristics of groups including species composition, body size, colour, sex, familiarity, parasite load and kinship (Krause and Ruxton, 2002). Female guppies actively prefer to associate with others of a similar size and those with which they have previously associated (Croft, 2003). Shoaling with kin in this context is likely to increase survival rate when facing high predation pressure because of the opportunity for sharing risks in a more cooperative way, for instance during predator inspection behaviour (Dugatkin, 1997). Pairs of fish that frequently engaged in predator inspection exchanged the front position more frequently which is likely to reduce the costs associated with this risky inspection behaviour (Croft et al., 2006). Wild female guppies showed a preference for particular partners within a highly interconnected social network of interactions, suggesting that co-operation networks might exist in free-ranging population of guppies (Croft et al., 2004, 2006). Recent work showed that there are assortative interactions on the basis of co-operative tendencies in social networks of guppies in the wild (Croft et al., 2009). However, the extent to which kinship plays a role in the formation of such networks is unknown partly because shoaling with kin could also be a by-product of phenotypic assortment. While our data show kin structure among shoals of juvenile guppies in some sites, we cannot, at present, distinguish between effects due to persistence of initial brood associations, active choice based on phenotype or active choice based on kin-recognition. Another intriguing question is whether the benefits of associating with kin change throughout ontogeny and whether the discrepancy between our work and Russell et al.'s (2004) study is due to the fact that kin-associations are more strongly selected for in juveniles.
The social organisation of animal groups is of great importance, influencing: choice of a sexual partner, development and maintenance of co-operative relationships, opportunities for social learning, and foraging and anti-predator behaviour. Our findings on kin assortment in guppies contribute to the understanding of the factors underlying shoal composition and highlight the potential of recent methodological advances in detecting such relationships.
References
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Arnold KE (2000). Kin recognition in rainbowfish (Melanotaenia eachamensis): sex, sibs and shoaling. Behav Ecol Sociobiol 48: 385–391.
Avise JC, Shapiro DY (1986). Evaluating kinship of newly settled juveniles within social-groups of the coral-reef fish Anthias squamipinnis. Evolution 40: 1051–1059.
Becher SA, Russell ST, Magurran AE (2002). Isolation and characterization of polymorphic microsatellites in the Trinidadian guppy (Poecilia reticulata). Mol Ecol Notes 2: 456–458.
Blouin MS (2003). DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol Evol 18: 503–511.
Breden F (2006). Guppies. Curr Biol 16: R865–R866.
Chapman BB, Morrell LJ, Benton TG, Krause J (2008). Early interactions with adults mediate the development of predator defences in guppies. Behav Ecol 19: 87–93.
Croft DP (2003). The social organisation of the guppy (Poecilia reticulata). PhD thesis. The University of Leeds: Leeds, UK.
Croft DP, Albanese B, Arrowsmith BJ, Botham M, Webster M, et al. (2003). Sex-biased movement in the guppy (Poecilia reticulata). Oecologia 137: 62–68.
Croft DP, James R, Thomas POR, Hathaway C, Mawdsley D, Laland KN et al. (2006). Social structure and co-operative interactions in a wild population of guppies (Poecilia reticulata). Behav Ecol Sociobiol 59: 644–650.
Croft DP, Krause J, Darden SK, Ramnarine IW, Faria JJ, James R (2009). Behavioural trait assortment in social networks: patterns and implications. Behav Ecol Sociobiol 63: 1495–1503.
Croft DP, Krause J, James R (2004). Social networks in the guppy (Poecilia reticulata). Biol Lett 271: 516–519.
Dugatkin LA (1997). Cooperation among animals: an evolutionary perspective. Oxford University Press: New York.
Emlen ST, Wrege PH (1988). The role of kinship in helping decisions among white-fronted bee-eaters. Behav Ecol Sociobiol 23: 305–315.
Endler JA (1980). Natural selection on color patterns in Poecilia reticulata. Evolution 34: 76–91.
Engel H, Kuppers C, Loffert D (2003). Highly efficient multiplex PCR using novel reaction chemistry. QIAGEN News 2: 41–43.
Evans JP, Kelley JL (2008). Implication of multiple mating for offspring relatedness and shoaling behaviour in juvenile guppies. Biol Lett 4: 623–626.
Fajen A, Breden F (1992). Mitochondrial DNA sequence variation among natural populations of the Trinidad guppy, Poecilia reticulata. Evolution 46: 1457–1465.
Foster KR, Wenseleers T, Ratnieks FLW (2006). Kin selection is the key to altruism. Trends Ecol Evol 21: 57–60.
Fraser DJ, Duchesne P, Bernatchez L (2005). Migratory charr schools exhibit population and kin associations beyond juvenile stages. Mol Ecol 14: 3133–3146.
Frommen JG, Bakker TCM (2004). Adult three-spined sticklebacks prefer to shoal with familiar kin. Behaviour 141: 1401–1409.
Frommen JG, Mehlis M, Brendler C, Bakker TCM (2007). Shoaling decisions in three-spined sticklebacks (Gasterosteus aculeatus)—familiarity, kinship and inbreeding. Behav Ecol Sociobiol 61: 533–539.
Gerlach G, Schardt U, Eckmann R, Meyer A (2001). Kin-structured subpopulations in Eurasian perch (Perca fluviatilis L.). Heredity 86: 213–221.
Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC (2007). High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. P Natl Acad Sci 104: 8913–8917.
Griffiths SW, Magurran AE (1997). Familiarity in schooling fish: how long does it take to acquire? Anim Behav 53: 945–949.
Griffiths SW, Magurran AE (1999). Schooling decisions in guppies (Poecilia reticulata) are based on familiarity rather than kin recognition by phenotype matching. Behav Ecol Sociobiol 45: 437–443.
Griffiths SW, Ward AJW (2006). Learned recognition of conspecifics. In: (Brown C, Laland KN, Krause J) (eds). Fish Cognition and Behavior. Blackwell Publishing: Oxford.
Hain TJA, Neff BD (2007). Multiple paternity and kin recognition mechanisms in a guppy population. Mol Ecol 16: 3938–3946.
Hamilton WD (1964). Genetical evolution of social behaviour. J Theor Biol 7: 1–52.
Hardy OJ, Vekemans X (2002). SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Herbinger CM, Doyle RW, Taggart CT, Lochmann SE, Brooker AL, Wright JM et al. (1997). Family relationship and effective population size in a natural cohort of Atlantic cod (Gadus morhua) larvae. Can J Fish Aquat Sci 54: 11–18.
Hiscock MJ, Brown JA (2000). Kin discrimination in juvenile brook trout (Salvelinus fontinalis) and the effect of odour concentration on kin preferences. Can J Zool 78: 278–282.
Houde AE (1997). Sex, color, and mate choice in guppies. Princeton University Press: New Jersey.
Kalinowski ST, Wagner AP, Taper ML (2006). ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6: 576–579.
Krause J, Butlin RK, Peuhkuri N, Pritchard VL (2000). The social organization of fish shoals: a test of the predictive power of laboratory experiments for the field. Biol Rev Camb Philos Soc 75: 477–501.
Krause J, Ruxton GD (2002). Living in groups. Oxford University Press: Oxford.
Ledesma JM, McRobert SP (2008). Shoaling in juvenile guppies: the effects of body size and shoal size. Behav Processes 77: 384–388.
Luikart G, England PR (1999). Statistical analysis of microsatellite DNA data. Trends Ecol Evol 14: 253–256.
Lynch M, Ritland K (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152: 1753–1766.
Magurran AE (2005). Evolutionary ecology: the Trinidadian guppy. Oxford University Press: London.
Magurran AE, Seghers BH (1990). Population differences in the schooling behavior of newborn guppies, Poecilia reticulata. Ethology 84: 334–342.
Mantel N (1967). Detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
Michalakis Y, Excoffier L (1996). A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142: 1061–1064.
Milinski M (1987). Tit-for-tat in sticklebacks and the evolution of cooperation. Nature 325: 433–435.
Naish KA, Carvalho GR, Pitcher TJ (1993). The genetic structure and microdistribution of shoals of Phoxinus phoxinus, the European minnow. J Fish Biol 43: 75–89.
Neff BD, Fu P, Gross MR (2000). Microsatellite multiplexing in fish. Trans Am Fish Soc 129: 584–593.
Nicholls JA, Double MC, Rowell DM, Magrath RD (2000). The evolution of cooperative and pair breeding in thornbills Acanthiza (Pardalotidae). J Avian Biol 31: 165–176.
Olendorf R, Reudi B, Hughes KA (2004). Primers for 12 polymorphic microsatellite DNA loci from the guppy (Poecilia reticulata). Mol Ecol Notes 4: 668–671.
Olsén KH (1999). Present knowledge of kin discrimination in salmonids. Genetica 104: 295–299.
Olsen KH, Petersson E, Ragnarsson B, Lundqvist H, Jarvi T (2004). Downstream migration in Atlantic salmon (Salmo salar) smolt sibling groups. Can J Fish Aquat Sci 61: 328–331.
Packer C, Gilbert DA, Pusey AE, O’Brien SJ (1991). A molecular genetic analysis of kinship and cooperation in African lions. Nature 351: 562–565.
Paradis E, Claude J, Strimmer K (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Paterson IG, Crispo E, Kinnison MT, Hendry AP, Bentzen P (2005). Characterization of tetranucleotide mlicrosatellite markers in guppy (Poecilia reticulata). Mol Ecol Notes 5: 269–271.
Peakall R, Smouse PE (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Peuhkuri N, Seppa P (1998). Do three-spined sticklebacks group with kin? Ann Zool Fenn 35: 21–27.
Pitcher TJ, Parrish JK (1993). Functions of shoaling behaviour in teleosts. Pages- 363–439. In T. J. Pitcher, editor. Behaviour of teleost fishes. Chapman & Hall, London.
Pouyaud L, Desmarais E, Chenuil A, Agnese TF, Bonhomme F (1999). Kin cohesiveness and possible inbreeding in the mouthbrooding tilapia Sarotherodon melanotheron (Pisces Cichlidae). Mol Ecol 8: 803–812.
Queller DC, Goodnight KF (1989). Estimating relatedness using genetic markers. Evolution 43: 258–275.
Raymond M, Rousset F (1995). Genepop (version-1.2)—population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Reznick DN, Butler MJ, Rodd FH, Ross P (1996). Life-history evolution in guppies (Poecilia reticulata) 6. Differential mortality as a mechanism for natural selection. Evolution 50: 1651–1660.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Rousset F (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145: 1219–1228.
Russell ST, Kelley JL, Graves JA, Magurran AE (2004). Kin structure and shoal composition dynamics in the guppy, Poecilia reticulata. Oikos 106: 520–526.
Wang J (2004). Sibship reconstruction from genetic data with typing errors. Genetics 166: 1963–1979.
Wang J (2007). Parentage and sibship exclusions: higher statistical power with more family members. Heredity 99: 205–217.
Wang J, Santure AW (2009). Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181: 1579–1594.
Ward AJW, Hart PJB (2003). The effects of kin and familiarity on interactions between fish. Fish Fish 4: 348–358.
Watanabe T, Yoshida M, Nakajima M, Taniguchi N (2003). Isolation and characterization of 43 microsatellite DNA markers for guppy (Poecilia reticulata). Mol Ecol Notes 3: 487–490.
Acknowledgements
Genotyping was performed at the Molecular Genetics Facility (Sheffield) supported by the Natural Environment Research Council (NERC), UK. We thank the SMGF staff; in particular Terry Burke, Deborah Dawson, Andy Krupa, Alain Frantz and Gavin Horsburgh for providing invaluable advice and assisting with genotyping and data analysis to CP. Thanks also go to Dominic Wright for his advice on how to collect tissue samples, Darren Croft for assisting in the field, Julien Claude for helping with R code, Indar Ramnarine for support with field equipment, Deborah Dawson and Vasilis Louca for comments on an early stage of the manuscript, and Jane Hughes, three anonymous reviewers, Pairot Pramual, Jolyon Dodgson and Tim Cushnie for greatly improving it at a later stage. Finally, CP and JK acknowledge financial support from the Thai Royal Government and the NERC (NE/D011035/1) respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Piyapong, C., Butlin, R., Faria, J. et al. Kin assortment in juvenile shoals in wild guppy populations. Heredity 106, 749–756 (2011). https://doi.org/10.1038/hdy.2010.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.115
Keywords
This article is cited by
-
Rhythm of relationships in a social fish over the course of a full year in the wild
Movement Ecology (2023)
-
Mating experiences with the same partner enhanced mating activities of naïve male medaka fish
Scientific Reports (2022)
-
How does host social behavior drive parasite non-selective evolution from the within-host to the landscape-scale?
Behavioral Ecology and Sociobiology (2021)
-
Neural pathways of olfactory kin imprinting and kin recognition in zebrafish
Cell and Tissue Research (2021)
-
Males optimally balance selfish and kin-selected strategies of sexual competition in the guppy
Nature Ecology & Evolution (2020)