Abstract
Genomic imprinting refers to the pattern of monoallelic parent-of-origin-dependent gene expression where one of the two alleles at a locus is expressed and the other silenced. Although some genes in mice are known to be imprinted, the true scope of imprinting and its impact on the genetic architecture of a wide range of morphometric traits is mostly unknown. We therefore searched for quantitative trait loci (QTL) exhibiting imprinting effects on mandible size and shape traits in a large F3 population of mice originating from an intercross of the LG/J (Large) and SM/J (Small) inbred strains. We discovered a total of 51 QTL affecting mandible size and shape, 6 of which exhibited differences between reciprocal heterozygotes, the usual signature of imprinting effects. However, our analysis showed that only one of these QTL (affecting mandible size) exhibited a pattern consistent with true imprinting effects, whereas reciprocal heterozygote differences in the other five all were due to maternal genetic effects. We concluded that genomic imprinting has a negligible effect on these specific morphometric traits, and that maternal genetic effects may account for many of the previously reported instances of apparent genomic imprinting.
Similar content being viewed by others
Introduction
Genomic imprinting occurs when the expression of a particular allele at a genetic locus depends on its parent of origin (Reik and Walter, 2001). Either the maternal or paternal allele may be expressed, with the alternate allele silenced by any of several epigenetic processes such as DNA methylation or histone modification (Lewis and Reik, 2006; Wood and Oakey, 2006; Allis et al., 2007). Increasing numbers of loci now are known to be imprinted, including a total of 154 in the mouse genome (Morison et al., 2005). However, more continue to be discovered (Luedi et al., 2005; Morison et al., 2005), and at present the actual total number of imprinted loci is unknown. The range of phenotypic effects produced by imprinted genes also is unclear (see Wolf et al., 2008), although these genes are known to affect growth and development (de Koning et al., 2000; Isles and Wilkinson, 2000) and cause several major human disorders such as the Prader–Willi and Angelman syndromes (Nicholls, 2000; Constancia et al., 2004; Heijmans et al., 2007).
The signature of parent-of-origin effects such as imprinting is a phenotypic difference between reciprocal heterozygotes, Aa and aA (where the paternal allele is listed first and the maternal allele second). However, Hager et al. (2008) have shown that genetic maternal effects can produce this same heterozygote difference and therefore be mistaken for imprinting. Genetic maternal effects occur when genes in the mother influence the environment (such as quantity and quality of her milk and overall maternal care), and thus the development, of her offspring (see Cheverud and Wolf, in press). Reciprocal heterozygote differences in the offspring of heterozygous mothers represent true imprinting effects, as these offspring all share the same maternal environment, whereas these differences in offspring born to homozygous (AA and aa) mothers are produced from genetic maternal effects (Hager et al., 2008).
Some recent genome scans for quantitative trait loci (QTL) have advanced our understanding of the relative impact of imprinting versus maternal effects on complex traits (Cheverud et al., 2008; Hager et al., 2008; Wolf et al., 2008). In an extensive study, Cheverud et al. (2008) discovered eight imprinted QTL affecting various internal organ weights in an F3 generation of mice produced from an original intercross of the LG/J (Large) and SM/J (Small) inbred strains. The imprinting effect was small, however; the average difference between reciprocal heterozygotes amounted to 0.25 standard deviations. In addition, reciprocal heterozygote differences in one QTL on chromosome 6 were due to genetic maternal effects (Cheverud et al., 2008). In the same mouse population, Wolf et al. (2008) found eight QTL with various imprinting patterns of effect on growth and weekly weights as well as five other QTL that exhibited maternal genetic (or a combination of maternal genetic and imprinting) effects on these traits (Hager et al., 2008).
Given these results for the growth and body composition traits in this F3 population (Cheverud et al., 2008; Hager et al., 2008; Wolf et al., 2008), we wanted to know to what extent genomic imprinting effects might contribute to the genetic architecture of complex morphological traits in these mice. We previously searched for QTL for mandibular size and shape in F2 mice from the Large × Small intercross (Klingenberg et al., 2001, 2004) and, therefore, decided to measure the same traits in the F3 mice to extend our additive/dominance effects model to include imprinting effects as well. We were particularly interested to discover whether some of the imprinted QTL uncovered by earlier analyses of size and growth traits (Cheverud et al., 2008; Hager et al., 2008; Wolf et al., 2008) would be the same as those for mandible size or shape, and to what extent parent-of-origin effects for these morphometric traits might turn out to be maternal genetic in origin.
Materials and methods
The population and traits
We made use of the F2 and F3 generations of mice reared from an original intercross of the LG/J and SM/J inbred strains (Kramer et al., 1998; Cheverud et al., 2008). Crossing of the F1 hybrids produced 510 F2 mice that in turn were crossed to yield 200 F3 families containing a total of 1632 mice. The F3 mice were cross-fostered by reciprocally exchanging half of the pups from pairs of litters born on the same day (Wolf et al., 2002). Altogether, 195 litters were involved in this cross-fostering regime. All mice from both generations were killed at 70 days of age (or after having reared their offspring to 3 weeks of age) at which time they were weighed and DNA extracted from their livers for subsequent genotyping. After killing, skeletonization was carried out by exposure to dermestid beetles.
A total of 353 polymorphic single-nucleotide polymorphism (SNP) markers were scored for all available F2 and F3 mice by the Illumina Golden-Gate assay (see Cheverud et al., 2008). These markers effectively covered all 19 autosomes, with an average interval of 4–5 cM between markers except for several regions in the genome where there was little polymorphism between the LG/J and SM/J strains (Hrbek et al., 2006). Haplotype reconstruction was accomplished with the PedPhase program (Li and Jiang, 2003a, 2003b) that allowed us to distinguish each of the four genotypes at each SNP locus in all F3 mice. These genotypes will be designated as LL, LS, SL and SS throughout the analysis that follows, where the paternal allele is listed first and the maternal allele second. The parent of origin of alleles could not be determined in the same manner for the F2 mice because their parents were genetically identical, and all analyses were based on mice in the F3 generation.
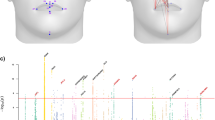
Both left and right sides of the mandibles in each mouse were separated at the mandibular symphysis, placed under a microscope, and their images were scanned into a computer. We then recorded x and y coordinates for 15 landmarks located around the outline of the mandible (see Figure 1). Once the first round of digitizing was accomplished, we performed a second round using the same images. Some mice had broken mandibles on one or both sides and thus were unusable, but where present, all mandibles had two replicate sets of coordinates. A total of 1889 mice were available for analysis, 374 from the F2 generation and 1515 from the F3 generation.
Size and shape traits
We used the x, y coordinate data to generate mandible size and shape measures for each mouse. For an overall measure of size, we computed centroid size by the square root of the sum of squared distances between each of the 15 landmarks and the centroid. The centroid, often referred to as the center of gravity, is that point at the means of the x and y coordinates of all landmarks (Dryden and Mardia, 1998). Centroid size was a one-dimensional trait measured in millimeters, and therefore was subjected to univariate statistical procedures throughout the analyses.
Shape, on the other hand, is an inherently multidimensional trait that involves all features of a configuration of landmark points except size, position and orientation. We used Procrustes superimposition (Dryden and Mardia, 1998) to generate a set of 30 new shape variables corresponding to the x and y coordinates at each of the 15 landmarks on the mandible that are independent of overall mandible size. The Procrustes procedure produces these shape variables by a four-step process of reflection, scaling, superimposition and rotation; see Klingenberg and McIntyre (1998) for a detailed description of the Procrustes procedure. This procedure eliminated four degrees of freedom in the mandible shape data, thereby reducing the number of dimensions of the resulting shape space from 30 to 26.
For each individual, we calculated the mean of the two replications for both left- and right-side measures (total of four values) for centroid size and each of the 30 Procrustes shape variables. Differences between sides (asymmetry) were not analyzed here and will be presented separately. The mean size and shape measures then were adjusted for sex, litter size and generation differences by obtaining residuals in an analysis of variance where these three factors were treated as classification variables. For centroid size, the mean for all F3 mice was 17.60 mm, with a standard deviation of 0.536.
QTL mapping
We searched for QTL for mandible centroid size and shape in the F3 mice using the regression approach of Haley and Knott (1992) implemented with the canonical correlation (CANCORR) procedure in SAS (SAS Institute, 1992) that previously has been described (Leamy et al., 1999; Klingenberg et al., 2001). To initiate this approach, we first assigned orthogonal genotypic index score values to each of the four possible genotypes at all SNP markers corresponding to additive (LL=+1, LS and SL=0, SS=−1), dominance (LS and SL=1, LL and SS=0) and imprinting or parent-of-origin-dependent (LS=+1, LL and SS=0, SL=−1) effects (see Mantey et al., 2005; Wolf et al., 2008). We then ran separate canonical correlation analyses of all three (additive, dominance, imprinting) index values with mandible centroid size, and with the mandible shape variables, at each marker on all 19 chromosomes. The probabilities associated with the F statistics generated in the canonical correlation analyses were logarithmically transformed (log of the probability (LPR)=log10(1/prob.)) to make the results comparable with logarithm of the odds (LOD) scores obtained by maximum likelihood analysis (Lander and Botstein, 1989). QTL were considered to be present at those sites of the highest LPR values that exceeded a specific threshold value.
To calculate threshold values, we performed separate permutation procedures (Churchill and Doerge, 1994) for mandible centroid size and mandible shape. Because individual mice in each of the 195 families in the F3 population were autocorrelated, however, we modified the usual permutation approach to ensure that the specific family structure in this generation was maintained. This was done by first calculating deviations of each individual from its family mean (for centroid size or the Procrustes shape variables) and randomly permuting these deviations within each family. We then randomly permuted all 195 family means and reconstructed new values for each individual by adding its permuted deviation to its new mean. After these new values for each individual were computed, we ran the canonical correlation analysis as before with the same additive, dominance and imprinting genotypic index scores for that individual and computed the highest LPR score on each chromosome. This procedure was done 1000 times, and chromosome-wise threshold values associated with the 5% probability level were obtained from the fifitieth highest values for each chromosome generated from these 1000 runs. To compute an experiment-wise (that is, genome-wise) threshold value, we identified the highest LPR value among all 19 chromosomes in each permutation run, and then calculated the fiftieth highest value from these LPR values (Churchill and Doerge, 1994).
If evidence for a QTL for mandible size or shape was found on a given chromosome, we tested for the presence of two QTL on that chromosome. To accomplish this, we ran canonical correlation analyses as before, but for all pairs of SNP locations on each chromosome. We computed χ2 values for both the 1- and 2-QTL models from Bartlett's V statistic (Green, 1978) and subtracted these two values to obtain the test statistic. If this difference exceeded the critical value for mandible size (distributed as χ2 with 3 degrees of freedom (size) or shape (78 degrees of freedom), then the 2-QTL model was considered the better fit and we concluded that two QTL were present on that chromosome. Again, because of the F3 family structure, however, we used the critical χ2 values obtained from the permutation analyses rather than conventional tabled values associated with these degrees of freedom.
We also tested for sex-specific QTL effects by first assigning a code for the sex of each F3 mouse. We then ran the canonical correlation analyses for the mandible size and shape traits to test for the interaction of sex with the additive, dominance and imprinting genotypic index values (partialing the main effects due to these three genotypic values and to sex). Chromosome-wise and genome-wise threshold LOD values were again estimated by permutation tests, and chromosomes with significant LOD values were assumed to contain a QTL whose effects differed in the two sexes. Where this occurred, we ran separate QTL analyses for males and for females.
QTL effects
Once the positions of all QTL on each chromosome affecting mandible centroid size were established, we estimated their additive (a), dominance (d), and imprinting (i) genotypic values. The a values estimate one-half of the difference between the average phenotypic values of the two homozygotes, the d values estimate the difference between the average phenotypic value of the heterozygotes and the midpoint between the two homozygote genotypic values (Falconer and Mackay, 1996), and the i values estimate one-half of the phenotypic difference between LS and SL heterozygotes (Wolf et al., 2008). Regression of mandible size on the additive, dominance and imprinting index values provided direct estimates of the a, d, and i values (and their standard errors). All genotypic values were divided by the standard deviation of centroid size to facilitate comparisons of their magnitudes among the QTL. To test these genotypic values for significance, we used the post hoc t-test results generated in the regression analyses, but with the threshold for significance determined by the mean of the 5% point-wise LPR values calculated for all 353 SNP sites (see Wolf et al., 2008).
For shape, we used multivariate regression for each QTL to estimate its additive, dominance and imprinting effects. This approach generated a, d and i vectors that had both a magnitude and a direction. To assess the overall magnitude of these effects for each QTL, we calculated the lengths of these vectors in units of Procrustes distance as follows: ∥a∥=(aTa)0.5, ∥d∥=(dTd)0.5, ∥i∥=(iTi)0.5. To depict these effects on mandibular shape, we graphed these changes with the use of thin-plate spline deformations (Bookstein, 1991; Dryden and Mardia, 1998) of the outline of the mandible. Although these graphs were useful in visualizing the shape changes produced by the QTL, it should be emphasized that they were constructed on the basis of the estimates of the effects at each landmark, and therefore provide no information about those sections of the mandible between landmarks.
For the QTL for mandible size or shape showing differences between reciprocal heterozygotes (significant i effects), we tested whether these differences were due to imprinting or to genetic maternal effects. This was accomplished using both of two different approaches suggested by Hager et al. (2008). In one approach, we used the same model as before that included additive, dominance and imprinting effects, but tested for the significance of i effects in offspring born to homozygous mothers (suggesting genetic maternal effects) versus heterozygous mothers (suggesting true imprinting). Where two QTL were indicated on the chromosome being tested, all a, d and i effects of the QTL not being tested were partialed out in the model. For the significance threshold, we used the mean of the 5% point-wise LPR values for i effects previously calculated.
As a second approach, we used a multiple regression model that included additive and dominance maternal genotypic scores obtained from F2 mothers as well as the usual additive, dominance and imprinting genotypic scores from all F3 progeny. In this model, a significant partial regression coefficient estimating i was taken as evidence of imprinting effects, whereas significant regressions for additive or dominance maternal values suggested genetic maternal effects. As before, we adjusted for the second QTL if two were present on a given chromosome, and assessed the significance of the imprinting or maternal additive/dominance effects using the mean of the 5% appropriate point-wise LPR values.
Results
Mandible centroid size
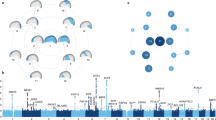
The results of the genome scan for QTL for mandible centroid size are given in Table 1. Each QTL is designated as CENT followed by its chromosome number and an extension that indicates whether it is the first or second QTL on that chromosome. Chromosomes 5 and 7 showed significant sex interaction effects in preliminary scans, but tests for QTL on both chromosomes did not reach significance for either males or females, so all QTL found were for the combined sexes. As may be seen, there are 14 different QTL for mandible size, 8 of which reached significance at the 5% genome-wise level (threshold LPR value=9.55) and the remaining 6 at the 5% chromosome-wise level (mean of the threshold LPR values=5.92). Two QTL are located on chromosome 2, whereas all others are on separate chromosomes. Their percentage of effect on mandible centroid size varies from 2.47 to 5.64% (CENT6.1), averaging 3.54%.
The additive genotypic values for the 14 centroid size QTL are generally about 0.2 standard deviations (mean of the absolute values=0.229). These values are significant for 13 of the QTL, and all these cases are positive in sign, suggesting that the LG/J alleles tend to increase mandible size. The mean of the (absolute) standardized dominance genotypic values is considerably less (0.069), this being reflected in the mean value of 0.34 for the (absolute) d/a ratio. In addition, only one QTL (CENT14.1) showed a significant dominance effect, so in general the QTL influencing centroid size exhibit primarily additive rather than dominance effects. The mean of the (absolute) i values for the 14 QTL is 0.081, slightly higher than the comparable value for dominance genotypic values, and four QTL show significant i effects. Using all significant a, d and i effects associated with the 14 QTL, a multiple regression analysis showed that these QTL jointly contribute 34% of the total variance in centroid size.
All four of the imprinted QTL exhibit negative i values, suggesting that SL heterozygotes are larger than LS heterozygotes. This trend can be seen in Figure 2 where the mean centroid size for each of the four genotypes at the SNP sites for these QTL are depicted. In addition, the figure shows the mean centroid size values for heterozygous (SL and LS) F3 mice born to homozygous and heterozygous mothers. For CENT2.1 and CENT6.1, it is clear that the SL/LS differences arise from offspring born to homozygous rather than heterozygous mothers. Both significance tests designed to distinguish imprinting from maternal genetic effects confirmed this trend and therefore these two QTL show genetic maternal rather than imprinting effects. For CENT3.1, there appears to be a difference between reciprocal heterozygotes born to heterozygous mothers, but again this difference did not reach significance at the point-wise threshold level for i effects (LPR=1.99<2.91; P>0.05), and the partial regression for maternal dominance effects was significant (LPR=2.32>2.07; P<0.05). This suggests that the overall SL/LS difference for this QTL once again is due to a genetic maternal effect.
Mandible size means (with bars indicating standard errors) in the F3 mice for each of the four genotypes of the QTL (CENT2.1, CENT3.1, CENT6.1 and CENT12.1) that exhibited significant apparent imprinting (i) effects. Means are also shown for heterozygous F3 mice (SL and LS) born to homozygous and heterozygous mothers.
For CENT12.1, reciprocal heterozygotes born from heterozygous parents show a significant difference (LPR=3.53, P<0.05), suggesting imprinting. Estimation of the i value from just these offspring yields a value of −0.190, somewhat less than that of −0.238 (Table 1) estimated from all offspring. Regression analysis also showed that these imprinting effects contributed 1.7% of the total variance in centroid size. Partial regression coefficients of maternal additive and dominance genotypic scores on mandible centroid size both were non-significant (P>0.05), confirming that this QTL exhibits imprinting effects and not a combination of imprinting and genetic maternal effects.
Mandible shape
Table 2 lists all mandible shape QTL (designated as SH) discovered in the genome scan. A total of 37 QTL reached significance, the LPR values for 33 of these exceeding the 5% genome-wise threshold value of 15.95. Chromosomes 16 and 19 each contain one QTL, whereas all remaining autosomes each contain two or more QTL. There are three QTL on chromosome 6, one affecting only females, another affecting only males and a third affecting both sexes. The lengths of the a vectors are significant for all QTL, varying from 0.21 to 0.56 and averaging 0.32. The lengths of the d vectors (mean=0.17) generally are less than those for the a vectors, although the mean ∥d∥/∥a∥ ratio is 0.57, somewhat higher than the comparable value of 0.34 for the mandible size QTL. Furthermore, 5 of the 36 QTL show significant dominance effects; so for mandible shape, dominance effects appear to be somewhat more important than for mandible size.
As regards potential imprinting effects, the average of the i vectors for all 36 shape QTL is 0.16, and these vectors are significant for just two QTL (SH7.1, and SH11.1). However, multivariate tests of the differences between the reciprocal heterozygous offspring produced from heterozygous mothers for these two QTL both were non-significant (P>0.05). These differences in offspring from homozygous mothers also were significant (P<0.05) in both cases, and the multivariate regression on maternal effects was significant (P<0.05) for SH7.1 (although not for SH12.1). These tests suggest that maternal rather than imprinting effects are acting at these two loci, and we therefore conclude that there is no evidence for genuine imprinting effects on mandible shape in this F3 mouse population.
Figure 3 illustrates the shape changes caused by the maternal (i) effects for these two QTL, with their additive and dominance effects shown as well for comparison. For both SH7.l and SH11.1, the changes seen generally are throughout the ascending ramus region. These displacements are quite subtle, however, and would not be discernible without appropriate scaling ( × 30). They are noticeably less than those for the dominance, and especially the additive, effects for these QTL.
Shape changes produced by two QTL (SH7.1 and SH11.1) with significant maternal genetic (i) effects on shape. For each QTL, the gray outline represents the shape for the overall mean configuration, whereas the dark line represents the mean shape due to additive (a), dominance (d) or maternal genetic (i) effects of these QTL. The QTL effects are quite subtle and have been multiplied by 30 for increased visualization.
Discussion
Impact of imprinting
We conducted this study to assess the impact of imprinting on mandible size and shape, and it was surprising to find few imprinting effects on these morphometric traits in the F3 mouse population. Although we did discover six QTL that showed statistically significant differences between reciprocal heterozygotes, for five of these QTL, the differences clearly were due to genetic maternal effects. Only one QTL on chromosome 12 exhibited significant imprinting effects (on mandible size), but by chance alone we would expect one of the 19 chromosomes to have a significant result for each of the two mandible traits. Therefore, we must conclude that imprinting has a negligible impact on the mandible traits in this F3 population of mice. This conclusion applies only to this specific population, however, and quite different results might be found in other populations that differ in their choice of the specific inbred progenitors or in the use of a different generation or numbers of individuals.
Our experimental design, especially including the large number of mice available in the F3 population, should have been optimal for the detection of imprinting effects on the morphometric traits. This population certainly was sufficient to enable Cheverud et al. (2008) and Wolf et al. (2008) to detect a number of imprinted QTL for various growth and body composition traits in the same mice. Cheverud et al. (2008) used a statistical model that tested for i effects singly by partialing a and d effects. We elected to follow the approach of Wolf et al. (2008) and to test for the joint effects of a, d and i and, if significant, then individually test each of these parameters for significance. However, preliminary statistical analyses using the model of Cheverud et al. (2008) showed significant i effects for just three QTL for mandible size and only one QTL for mandible shape. If anything, therefore, our statistical model was more liberal in detecting i effects in the QTL for mandible size and shape. Power analyses of the similar model presented in Wolf et al. (2008) confirms this assertion, demonstrating that the approach used herein to detect loci is generally powerful and robust.
QTL effects on mandible size and shape
We had ample opportunity to test for imprinting effects because our analysis uncovered an abundance of QTL, 14 for mandible size and 37 for mandible shape. This is an increase over the 12 QTL for mandible size and especially the 25 QTL for mandible shape we previously found in an original study using the F2 generation of mice (Klingenberg et al., 2001). No doubt this increase was partly due to the much larger number of mice available in the F3 generation. In addition, however, we digitized 15 landmark points in the F3 mice compared with just 5 in the previous study (Klingenberg et al., 2001), and we fully expected to find additional QTL affecting the more complex multidimensional shape trait that was produced.
In another study making use of the F2 mice, Klingenberg et al. (2004) used 16 landmark points to test for integration and modularity of QTL effects on mandible shape. These points included the same 15 used in this study as well as an additional point on the condylar process (Klingenberg et al., 2004; Figure 1). Klingenberg et al. (2004) discovered a total of 33 shape QTL, and inspection of their locations suggests that 27 (81%) of these are replicated in this study. The remaining 10 of our 37 shape QTL (Table 2) appear to be at different locations in the genome, including two QTL on chromosome 3 where none were found in the previous study. Although the true level of QTL replication is difficult to estimate because of the experimental error inherent in mapping, we would expect the locations of the shape QTL we have discovered here to be more accurate than those previously estimated because of the additional round of recombination in the F3 generation and the larger number of mice we used.
The QTL for mandible size we discovered exhibited overwhelmingly additive effects, 13 of the 14 showing significant a values. Only one QTL, CENT14.1, showed significant dominance genotypic effects, although the magnitude of this effect (d value) was less than its additive effect (a). Klingenberg et al. (2001) also found that only one QTL (chromosome 4) of 12 affecting mandible centroid size in the F2 generation of the Large × Small intercross mice showed significant dominance effects (d=0.072, a=0.036). Overall, therefore, dominance does not appear to be very important for these QTL for mandible size, although it should be noted that the statistical power to detect additive effects is greater than that for dominance effects. Four QTL showed significant i effects, suggesting that reciprocal heterozygote differences were somewhat more prominent (or perhaps more detectable) than dominance effects for these QTL.
Additive effects also were quite important for the QTL affecting mandible shape, the length of the a vectors being significant in all 37 instances. Five QTL showed significant dominance effects, as did 6 (of 33) QTL for mandible shape in the F2 mice (Klingenberg et al., 2004). Interestingly, the five QTL showing dominance are located on entirely different chromosomes than the six found by Klingenberg et al. (2004). It is difficult to assess repeatability of this sort, however, as this may be a consequence of including i effects in our model rather than only a and d as were used in the previous study. Only two QTL affecting mandible shape exhibited significant i values, so parent-of-origin effects appear to be even less important for the QTL affecting mandible shape. Among all 51 total QTL affecting both mandible size and shape, in general, the frequency of significant i effects (differences between reciprocal heterozygotes) is comparable with that found for dominance effects. This also was the case for the QTL discovered for various body composition traits in these mice (Cheverud et al., 2008).
Parent-of-origin patterns
Wolf et al. (2008) have classified patterns of imprinting into three basic categories: parental (including either paternal or maternal) expression, polar dominance imprinting and bipolar dominance imprinting (see also Cheverud et al., 2008). Parental imprinting (that is, maternal or paternal expression) is characterized by each of the heterozygotes resembling the homozygote sharing the same parent of origin of the expressed allele, polar dominance imprinting by one of the heterozygotes being larger or smaller than all other genotypes and bipolar dominance imprinting by one heterozygote being larger and the other smaller from the other two genotypes that are equal. For CENT12.1, the one QTL that exhibited true imprinting effects, the mandible size means for LL, LS, SL and SS offspring born to heterozygous mothers are 17.49, 17.48, 17.68 and 17.63, yielding standardized a, d and i values of −0.13, +0.03 and −0.19, respectively. This suggests that this QTL exhibits parental, imprinting, and specifically because the a/i ratio is positive, paternal expression (Wolf et al., 2008). In their analysis of imprinted QTL affecting various body composition traits in these mice, Cheverud et al. (2008) found 13, 11 and 4 instances of parental expression, bipolar dominance imprinting and polar dominance imprinting, respectively. In an analysis of body size and growth traits, Wolf et al. (2008) found four loci showing paternal expression, one showing bipolar dominance and one showing only polar overdominance, with the four other loci showing patterns that changed over ontogeny.
The identity of CENT12.1 is unknown and in fact occurs in a region (20.20 cM on chromosome 12) where no imprinted QTL have been reported in mice (Morison et al., 2005). However, Luedi et al. (2005) developed a statistical model on the basis of DNA sequence characteristics that predicted 600 different murine genes that may be imprinted, including several in the confidence interval for CENT12.1. Two of these are Foxg1 (21 cM) and Twist1 (16.0 cM), both of which produce various craniofacial effects (Eppig et al., 2005) and that are predicted to exhibit a paternal imprinting pattern of expression (Luedi et al., 2005). Foxg1 was also listed as a candidate gene for an imprinted QTL previously discovered on chromosome 12 (at 30.56 cM) that had strong effects on reproductive fat pad, liver, kidney and total body weight in this same population of F3 mice (Cheverud et al., 2008).
It was interesting that the other three QTL with apparent parent-of-origin effects on centroid size mirrored the pattern for CENT12.1 where LS heterozygotes were smaller than SL heterozygotes. Yet these differences between reciprocal heterozygotes, especially for CENT2.1 and CENT6.1 (see Figure 2), were generated almost entirely in the offspring from homozygous mothers, making them clearly maternal genetic in origin. This was also true for the two QTL with significant i effects on mandible shape.
Our findings clearly suggest caution in assuming that parent-of-origin patterns in QTL studies are solely because of imprinting. Imprinting effects can be tested without the confounding influence of maternal effects in QTL analyses that make use of the F2 generation from an original intercross of inbred strains, as in this case the F1 parents are all genetically identical (Hager et al., 2008). This also is the case for crosses from outbred lines if it is assumed that the QTL are fixed for alternative alleles between lines (de Konig et al., 2002). Where this assumption cannot be made, however, as in advanced intercross lines such as that used in this investigation, it is important to distinguish between imprinting versus genetic maternal effects. As more experiments are done where data are available to make this distinction, it will be interesting to discover the proportion of putative imprinting genes that might really be exerting their effects through the maternal environment.
Conclusion
We discovered a large number of QTL affecting mandible size and shape in our F3 mouse population, but only a single QTL exhibited true imprinting effects. Whether this result might generally be found for other sorts of skeletal traits cannot be known without further studies. It is suggestive, however, that Cheverud et al. (2008) found a number of imprinted QTL for five organ weights and overall body weight in these mice, but none for tail length. On the other hand, the Imprinted Gene Catalog (Morison et al., 2005) contains at least five imprinted genes affecting the mouse skeleton and no doubt more will be discovered in time. Only when we have a more comprehensive knowledge of the number and effect of imprinted QTL for morphometric traits such as those used here we can fully assess their contribution to the genetic architecture of these traits.
References
Allis CD, Jenuwein T, Reinberg D (2007). Epigenetics. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York.
Bookstein FL (1991). Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge University Press: Cambridge, UK.
Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB (2008). Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci 105: 4253–4258.
Cheverud JM, Wolf JB (in press). Genetics and evolutionary consequences of maternal effects. In: Maestripieri D, Mateo JM (eds). Maternal Effects in Mammals. University of Chicago Press: Chicago.
Churchill GA, Doerge RW (1994). Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971.
Constancia M, Kelsey G, Reik W (2004). Resourceful imprinting. Nature 432: 53–57.
de Konig DJ, Bovenhuis H, van Arendonk AM (2002). On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics 161: 931–938.
de Koning DJ, Rattink AP, Harlizius B, Van Arendonk JAM, Brascamp EW, Groenen MAM (2000). Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc Natl Acad Sci USA 97: 7947–7950.
Dryden IL, Mardia KV (1998). Statistical Analysis of Shape. Wiley: Chichester, UK.
Eppig JT, Bult CJ, Kadin JA, Richardson JE, Blake JA, and the members of the Mouse Genome Database Group (2005). The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res 33: D471–D475.
Falconer DS, Mackay TFC (1996). Introduction to Quantitative Genetics, 4th edn, Longman: Essex, UK.
Green PE (1978). Analyzing Multivariate Data. Dryden: Hinsdale, Illinois.
Hager R, Cheverud JM, Wolf JB (2008). Maternal effects as the cause of apparent parent-of-origin dependent effects that mimic genomic imprinting. Genetics 178: 1755–1762.
Haley CS, Knott SA (1992). A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324.
Heijmans BT, Kremer K, Tobi EW, Boomsma EI, Slagboom PE (2007). Heritable rather than age-related environmental stochastic factors dominate variation in DNA methylation of the human IGF2/H10 locus. Hum Mol Genet 16: 547–554.
Hrbek TR, de Brito BA, Wang B, Pletscher LS, Cheverud JM (2006). Genetic characterization of a new set of recombinant inbred lines (LG × SM) formed from the intercross of SM/J and LG/J inbred mouse strains. Mamm Genome 17: 417–429.
Isles AR, Wilkinson LS (2000). Imprinted genes, cognition and behaviour. Trends Cogn Sci 4: 309–318.
Klingenberg CP, Leamy LJ, Cheverud JM (2004). Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics 166: 1909–1921.
Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM (2001). Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157: 785–802.
Klingenberg CP, McIntyre GS (1998). Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution 52: 1363–1375.
Kramer MG, Vaughn TT, Pletsher LS, King-Ellison K, Adams E, Erikson C et al. (1998). Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet Mol Biol 21: 211–218.
Lander ES, Botstein D (1989). Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199.
Leamy LJ, Routman EJ, Cheverud JM (1999). Quantitative trait loci for early- and late-development skull characters in mice: a test of the genetic independence model of morphological integration. Am Nat 153: 201–214.
Lewis A, Reik W (2006). How imprinting centres work. Cytogenet Genome Res 113: 81–89.
Li J, Jiang T (2003a). Efficient rule-based haplotyping algorithms for pedigree data. Proc Recomb 3: r291–r294.
Li J, Jiang T (2003b). Efficient inference of haplotypes from genotypes on a pedigree. J Bioinform Comput Biol 1: 41–69.
Luedi PP, Hartemink AJ, Jirtle RL (2005). Genome-wide prediction of imprinted murine genes. Genome Res 15: 875–884.
Mantey C, Brockmann GA, Kalm E, Reinsch N (2005). Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered 96: 329–338.
Morison IM, Ramsay JP, Spencer HG (2005). A census of mammalian imprinting. Trends Genet 21: 457–465.
Nicholls RD (2000). The impact of genomic imprinting for neurobehavioral and developmental disorders. J Clin Investig 105: 413–418.
Reik W, Walter J (2001). Genomic imprinting: parental influence on the genome. Nat Rev Genet 2: 21–32.
SAS Institute Inc (1992). SAS® Technical Report, SAS/STAT Software: Changes and Enhancements, Release 6.07. SAS Institute Inc.: Cary, NC.
Wolf JB, Cheverud JM, Roseman CC, Hager R (2008). Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet 4: e1000091.
Wolf JB, Vaughn TT, Pletscher LS, Cheverud JM (2002). Contribution of maternal effect QTL to genetic architecture of early growth in mice. Heredity 89: 300–310.
Wood AJ, Oakey RJ (2006). Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet 2: 1677–1685.
Acknowledgements
We thank S Sen for suggesting the approach used for calculating threshold LOD scores and three anonymous reviewers for useful revision suggestions. This research was supported by grants from the Biotechnology and Biological Sciences Research Council, UK (BBSRC), an Underwood Fellowship from the BBSRC and NIH Grants DK055736 and RR015116 and NSF BCS-0725068.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leamy, L., Klingenberg, C., Sherratt, E. et al. A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity 101, 518–526 (2008). https://doi.org/10.1038/hdy.2008.79
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.79
Keywords
This article is cited by
-
Genetic analysis of the mandible morphology in DDD.Cg-\(A^{y}\)/Sgn and C57BL/6J inbred mice
Journal of Genetics (2019)
-
Performance of single and multi-atlas based automated landmarking methods compared to expert annotations in volumetric microCT datasets of mouse mandibles
Frontiers in Zoology (2015)
-
Using whole-genome sequences of the LG/J and SM/J inbred mouse strains to prioritize quantitative trait genes and nucleotides
BMC Genomics (2015)
-
Effect of chromosomal reorganizations on morphological covariation of the mouse mandible: insights from a Robertsonian system of Mus musculus domesticus
Frontiers in Zoology (2014)
-
Body composition and gene expression QTL mapping in mice reveals imprinting and interaction effects
BMC Genetics (2013)