Abstract
The olive fruit fly, Bactrocera oleae, is the main pest of the olive fruit and its expansion is exclusively restricted to the cultivation zone of the olive tree. Even though olive production has a century-old history in California, the olive fly was first detected in the Los Angeles area in 1998. Within 5 years of the first observation, the insect was reported from all olive cultivation areas of the state. Field-collected flies from five locations in California and another from Israel were analyzed on the basis of microsatellite polymorphisms in 10 microsatellite loci. These results were integrated with those of a previous study of olive fly populations around the European part of the Mediterranean basin. The analysis pointed to the eastern part of the Mediterranean as the putative source of the observed invasion. Moreover, samples from California were quite different from Mediterranean samples implying the participation of phenomena such as genetic drift during the invasion and expansion of the olive fly in California.
Similar content being viewed by others
Introduction
The olive fly, Bactrocera oleae (Gmelin), is a member of the Diptera family Tephritidae. Being the most important pest of cultivated olives, its management is a matter of great importance. The olive fly damages the fruit by ‘stings’ resulting from oviposition. The hatched larva feeds on the mesocarp of the olive, creating channels inside the fruit. The oviposition site may also serve as a point of entry for microbial infection of the fruit, compromising the organoleptic qualities of the olive. This may result in reduced commercial value of table olives and unmarketable olive oil because of high levels of acidity. These factors can result in an annual production loss of between 5 and 30%, depending on environmental conditions (Mazomenos, 1989; Katsoyannos, 1992).
Being a monophagus insect, the range of the olive fly is limited to the olive cultivation zones, as well as areas where olive trees are indigenous. Traditionally, this was restricted to the Mediterranean basin where most olive trees are grown. However, olive cultivation is expanding in areas such as South Africa, Australia, China and the Americas.
The Spanish first carried olive tree cuttings to the Americas in the middle of the sixteenth century. Around 1700, Franciscan monks brought olive trees to Mexico and from there to California. Intensive cultivation of the olive tree started in central and southern California at the end of the nineteenth century. Today the California olive industry produces table and oil olives valued at about US$80 million (California Department of Food and Agriculture, 2006). The olive fly was first found in California near Los Angeles International Airport in October 1998 (Rice, 2000). A statewide monitoring program subsequently detected the olive fly in all olive cultivation areas of the state over the following 5 years (Zalom et al., 2008). However, the geographical origin of the insect's invasion remains unknown.
Analyses of natural olive fly populations support the subdivision into three groups: Pakistan, Africa and Mediterranean plus America (Nardi et al., 2005), with the Mediterranean population further differentiated into eastern (Cypriot), central (Greek–Italian) and western (Iberian) groups (Augustinos et al., 2005). The fundamental aim of the present study was a precise determination of the geographical origin of the California invasion of the olive fly within the Mediterranean group. Field-collected samples from five different locations of California and one from Israel were analyzed on the basis of polymorphism of 10 microsatellite loci. These markers constitute a subset of the markers described and used in the analysis of endemic olive fly populations around the Mediterranean basin by Augustinos et al. (2002, 2005). Microsatellites are particularly informative in the study of recent population phenomena, such as biological invasions (Bruford and Wayne, 1993; Schlötterer and Pemberton, 1994; Tautz and Schlötterer, 1994). Moreover, they have been successfully used in the analysis of population structure of different Tephritidae species, such as Ceratitis capitata (Bonizzoni et al., 2001; Gasperi et al., 2002), Bactrocera dorsalis (Aketarawong et al., 2007) and B. tryoni (Yu et al., 2001; Gilchrist et al., 2004). In the case of C. capitata, they have provided guidance in establishing the source of recent invasions in different parts of the world (Meixner et al., 2002; Bonizzoni et al., 2004). Such analyses can contribute to better planning of control strategies to avoid future infestations, as a detailed knowledge of the biology, genetic structure and geographical variability of a given species is a prerequisite to establishing control measures such as quarantine, phytosanitary control and eradication (Roderick and Navajas, 2003).
Materials and methods
Collection of fly samples and DNA extraction
Collection sites and number of flies used in the study are shown in Figure 1 and Table 1. Individual flies were collected during the olive-harvesting season from infested olive fruit. Flies were stored either at −80 °C or in 95% ethanol at −20 °C. Genomic DNA was extracted using the Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA).
Collection sites and geographical representation of the clustering outcome, assuming three hypothetical clusters. (a) Map showing the collection sites in the Mediterranean basin. 1, Gimmaraes-PO; 2, Lisbon-PO; 3, Murcia-SP; 4, Madrid-SP; 5, Arrhenys-SP; 6, Farfa-IT; 7, Vasto-IT; 8, Alexandroupolis-GR; 9, Lefkas-GR; 10, Patras-GR; 11, Mani-GR; 12, Ithaca-GR; 13, Kos-GR; 14, Kythira-GR; 15, Maladrino-GR; 16, Crete-GR; 17, Aidin-TU; 18, Nicosia-CY; 19, Limassol-CY; 20, Sde Boker-IS. TU, Turkey; IS, Israel; CY, Cyprus; GR, Greece; IT, Italy; SP, Spain; PO, Portugal. (b) Map showing the collection sites in California. 21, Calaveras; 22, Napa; 23, Solano; 24, Yolo Davis; 25, San Luis Obispo. Colored components in each pie show the co-ancestry distribution of individuals in each one of the three clusters. The colour reproduction of this figure is available on the html full text version of the manuscript.
Genotyping
We used 10 microsatellite markers in the present analysis, named Boms2, 10, 18, 21, 22, 25, 29, 30, 31 and 61 (Augustinos et al. 2002, 2005). PCRs were performed in a total volume of 10 μl, containing 10 ng of genomic DNA, 1 × complete reaction buffer (New England Biolabs, Inc., Ipswich, MA, USA), 0.2 mM of each dNTP, 0.5 μM of each primer and 0.4 U of Taq polymerase (New England Biolabs, Inc.). Amplifications were performed with an initial denaturation step (5 min at 95 °C), followed by 30 cycles of: 15 s at 94 °C, 30 s at 50 °C and 30 s at 72 °C, with a final elongation step of 5 min at 72 °C. PCR products were electrophoresed on 5% denaturing polyacrylamide gels and visualized according to the Silver Sequence DNA Sequencing System technical manual (Promega).
Data analysis
Genetic variability was measured as the mean number of alleles per locus (na), effective number of alleles (ne), observed (Ho) and expected heterozygosity (He), using POPGENE version 1.31 (Yeh et al., 1999) and allelic richness after correction for sample size, using FSTAT (Goudet, 2001). Deviations from Hardy–Weinberg equilibrium (HWE) were tested with the G2 likelihood ratio test in POPGENE. G2 criterion [defined as (Observed value) × ln(Observed value/Expected value)]. Genotypic disequilibrium was tested with Genepop (Raymond and Rousset, 1995), with Fisher's exact test, for all pairs of loci in all samples and across samples.
Genetic distances were measured according to Nei (1972) using POPGENE (Yeh et al., 1999). A UPGMA dendrogram was constructed in PHYLIP 3.6C (Felsenstein, 1994), using the allele frequencies. The robustness of each node was assessed by the bootstrap method. For this purpose, 100 pseudoreplicates were generated by random resampling of the original data, using SEQBOOT and GENDIST. The new distance matrices were then subjected to NEIGHBOR and CONSENSE to produce a consensus tree. Visualization of the dendrogram was performed by TreeView32 software (Page, 1996).
Pairwise FST values were estimated using FSTAT software. Inference on the degree of population subdivision based on hierarchical analysis of molecular variance (AMOVA) was performed by the ARLEQUIN 2.0 software (Schneider et al., 1997), to compare the percentage of genetic variability attributed to the variance among the major geographical areas sampled to that observed among samples within each of them. The significance of the resultant F statistics and variance components were tested with 10 000 permutations.
STRUCTURE software was used to determine the number of possible genetic clusters in California (Pritchard et al., 2000; Falush et al., 2003). We used all four different models, with a burn-in period of 50 000 and 50 000 Markov chain Monte Carlo (MCMC) repetitions after the initial burn-in. We tested for K=1 to K=5 and performed five runs of each model, to check the consistency of the results. In a previous study we observed at least three subpopulations of the olive fly in the Mediterranean, but these were not well differentiated due to gene flow. We used again the four different models, with a burn-in period of 50 000 and 50 000 MCMC repetitions after the initial burn-in, to analyze the structuring of all 25 samples. We ran five repetitions of all above models (to check the consistency of our results), assuming K=1 to K=10. Although the authors of the program recommend the use of the admixture model, they also suggest that the no-admixture model is more powerful at detecting subtle structure, such as in our case. Because the likelihood curve did not provide a clear K-value (Figure 3a), we estimated a modified K-value, taking also into account the consistency of the results. On the basis of modification described by Evanno et al. (2005) we divided the mean likelihood value of the five runs by their standard deviation and considered this as the modified new K-curve (Figure 3b).
GENALEX 6.1 software (Peakall and Smouse, 2006) was used to estimate pairwise population PhiPT values and PhiPT matrix was used to perform principal components analysis (PCA). PhiPT measure suppresses within-population variance and simply calculates population differentiation based on the genotypic variance.
GeneClass 2.0 software (Piry et al., 2004) was used to perform population assignment and exclusion test and to calculate the probability of origin for each individual and each sample. We also tried to assign California samples, assuming four subpopulations in the Mediterranean basin (the three previously described along with the Israeli). We chose a Bayesian model using the Rannala and Mountain (1997) criterion, under a 0.05 threshold.
Finally, we used BOTTLENECK software (Cornuet and Luikart, 1997) to detect any recent bottleneck phenomena, especially in California. We chose the Wilcoxon sign-rank test because it performs better than the other two tests with few loci.
Results
The Mediterranean basin is presumably the source of the olive fly invasion in California (Nardi et al., 2005). To further specify the origin of this invasion, microsatellite polymorphism data of California samples were compared with the preexisting data set of Mediterranean samples (Augustinos et al., 2005). To expand the Mediterranean data set, a sample from Israel was also included in the analysis. For this purpose, 150 field-collected flies from five different locations in California (Calaveras, Napa, Yolo, Solano and San Luis Obispo) and 18 from Israel (Sde Boker), as shown in Figure 1 and Table 1, were genotyped at 10 microsatellite loci.
Hardy–Weinberg equilibrium and linkage disequilibrium
Conformation to HWE was tested in California and Israeli samples according to the G2 criterion at a significance level of 5%. Out of 60 tests performed (6 samples × 10 loci), none of them showed departure from HWE after the sequential Bonferroni correction (Rice, 1989). Interestingly, testing HWE under the assumption of a single California population, two loci deviated: Boms25 due to homogeneity excess and Boms29 due to an excess of one class of homozygotes. This contrasts the situation in the Mediterranean basin where no deviations were observed (Augustinos et al., 2005) and may evidence the fact that there is not a single, homogeneous population of the olive fly in California. This could be the result of the very rapid expansion of the species in a large geographical area, where genetic drift and subsequent allele adaptation in particular ecological niches took place faster than the homogenizing effects of unrestricted gene flow. Linkage disequilibrium was observed only in few of the tests performed. However, after correcting for multiple tests, no linkage disequilibrium was statistically significant.
Microsatellite variability in California and Israel
Genetic variability was measured as the average number of actual (na) and effective alleles (ne), allelic richness, and degree of observed (Ho) and expected (He) heterozygosity (Table 2). A westward decline in the levels of polymorphism (He values) in the olive fly Mediterranean populations has already been reported (Augustinos et al., 2005). The trend holds for the Israeli sample (He=0.59), whose value is greater than those of the Central Med samples, even though it is a small sample and estimation errors cannot be excluded. California ne, Ho and He values are intermediate between those observed in the West Med and the Central Med groups. Allelic richness in California is the lowest among all regions, which is in accordance with the hypothesis of a Mediterranean source of olive fly invasion in California (Nardi et al., 2005).
Analysis of alleles gave one new allele in California, albeit at very low frequency. In addition, three of the California alleles were not sampled in the Iberian Peninsula. Given the large number of flies collected in Spain and Portugal (168), it is highly unlikely that these three alleles are present in this area. Moreover, no private alleles were found among West Med and California, West Med, East Med and California, and among West and East Med. This indicates that the West Med samples are quite discrete from the East Med and California samples.
Population structure in California
Estimated pairwise FST values in California were all not significant except for one (Sol-CA/SLO-CA), as shown in Table 3. Furthermore, all FST values among California samples and Mediterranean samples are significant (except those of Aid-TU), although at different probability levels. Genetic distances among California samples were measured according to Nei (1972 ; Table 3) and these varied between 0.0124 (Sol-CA/Yol-CA) and 0.0450 (SLO-CA/Sol-CA).
Further analysis of olive fly population structure in California was performed using the STRUCTURE software. Five independent runs of all four models were performed, assuming K ranging between 1 and 5. There seems to be no clustering of California samples. To further analyze the genetic structure, pairwise PhiPT values were calculated and PhiPT pairwise matrix was used to perform PCA. Still, no clustering of samples inside California was observed.
Worldwide structuring of olive fly samples
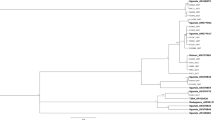
Analysis of significance of pairwise FST values among all samples showed the possibility of clustering in four subpopulations (Table 3). West Med samples are significantly different from all other samples. Similarly, California samples constitute a separate group, as they are significantly different from all other samples. Central Med samples form yet another group, whereas Cypriot and Israeli samples seem to comprise a further, East Med cluster. Aid-TU sample is very small (nine individuals) and this is probably the reason it presents no significant difference with almost any other sample. Genetic distances among California and all the other samples ranged between 0.0459 (Yol-CA/Man-GR) and 0.1975 (SLO-CA/Mad-SP). The Israeli sample (Sde-IS) showed a minimum genetic distance with Lim-CY (0.0496) and a maximum genetic distance with Mad-SP (0.1459). These values were used to construct an unrooted UPGMA dendrogram (Figure 2). The topology of the dendrogram was confirmed by relatively high bootstrap values at the main nodes (100 and 78%). The major clusters as well as further groupings within these clusters were identical with those of the Augustinos et al. (2005) analysis, albeit generated with 10 (of the original 12) microsatellite loci. There are the three previously described subpopulations of Western Mediterranean (Spain and Portugal), Central Mediterranean (Greece, Italy and Turkey) and Eastern Mediterranean (Cyprus). Interestingly, samples from Israel and California fall in this third cluster. These results point to the eastern part of the Mediterranean basin as a putative source of the olive fly invasion in California.
UPGMA dendrogram constructed after 100 bootstrap resamples, based on Nei's (1972) genetic distances, showing the relationships among the 25 samples studied. TU, Turkey; IS, Israel; CY, Cyprus; GR, Greece; IT, Italy; SP, Spain; PO, Portugal; CA, California.
STRUCTURE analysis was performed, assuming K ranging between 1 and 10. The no-admixture model provided a better clustering of our samples. Although we expect our samples to be admixed, the no-admixture model produces better results when genetic structure is not very clear due to high gene flow (as in our case). It seems that there are at least three subpopulations, but the likelihood curve is not clear as to whether K can be greater than 3 (Figure 3a). K-curve continues growing after the value of 3, presumably beyond the true K value. However, independent runs are not so consistent, especially when assumed K grows. To incorporate consistency to our results and based on the modification described by Evanno et al. (2005), we estimated a new K-curve by dividing the mean likelihood of five independent runs by their standard deviation (Figure 3b). This way it becomes apparent that the true number of clusters must be three and, surely, no more than four. Partition of color components in Figure 4 (K=3) substantially differentiates West Med (1–5) from East Med flies (18–20), whereas Central Med flies fall into a ‘transition zone’ that seems to share a great degree of ancestry either with Iberian or with Cypriot–Israeli populations. However, California flies (21–25) are clearly more alike those of the East Med. Table 4 presents the average number of ancestry probabilities of each one of the 25 samples in each of the three hypothetical clusters, whereas Figure 1 shows how the assumed three genetic groups are distributed among the 25 samples. It is obvious that individuals of each sample are distributed to all three classes, probably due to the high gene flow between olive fly natural populations in the Mediterranean basin.
STRUCTURE analysis: mean variance of likelihood for all samples, assuming K=1 to K=10 under the no-admixture with correlated allele frequencies model. Each value represents the mean of five independent runs. (a) Estimation of the true number of clusters, based on the likelihood (K) curve. (b) Estimation of the true number of clusters, based on the likelihood (K) curve, taking into account consistency of the results. The colour reproduction of this figure is available on the html full text version of the manuscript.
STRUCTURE analysis: estimated population structure for all samples based on allele frequency variation. Each individual is represented by a vertical line which is partitioned into K=3 colored components. 1–2, Portugal; 3–5, Spain; 6–7, Italy; 8–16, Greece; 17–18, Cyprus; 19, Turkey; 20, Israel; 21–25, California. The colour reproduction of this figure is available on the html full text version of the manuscript.
To further analyze structuring of olive fly populations, PCA was performed with the GENALEX software. Initially, PhiPT values were estimated and the PhiPT pairwise matrix was used to perform PCA. This analysis clearly demonstrates that there are at least four clusters: the first is formed by West Med samples (Iberian Peninsula), the second by Central Med samples (Greece, Italy and Turkey), the third by East Med samples (Cyprus and Israel) and the fourth by California samples (Figure 5). However, it is clear that California samples are most closely related to the East Med group and most distantly to the West Med group.
Analysis of molecular variance
AMOVA was performed to test the degree of homogeneity among populations. Samples were grouped in four populations, based on geographical criteria (West Med, Central Med, East Med and California) and on PCA results. Most variation is attributed to within-sample variability (95.6% of the total variance, FST=0.04397). However, a substantial proportion is due to differences among groups (3.79%). Only a very small, nonsignificant proportion of the variance derives from within-group variation (0.6%).
Migration
GeneClass version 2.0 was used to assign all individuals to their respective samples and 373 of the 840 individuals were correctly identified (44.4%). When all Mediterranean samples were clustered into five subpopulations (West Med, Central Med, Cypriot, Israeli and California), 651 of the 840 individuals (77.5%) were correctly assigned (Appendix 1). It should be noted that for this analysis we divided East Med cluster into Cypriot and Israeli, in an effort to identify more precisely the source of the California invasion. West Med individuals were properly assigned as such at 82.1%, whereas 12.5% were assigned as Central Med, very few as Israeli and Californian, and none as Cypriot. Individuals from the Central Med subpopulation presented the lowest degree of correct assignment (34.7% for respective samples—60.2% for the Central Med cluster), illustrating the extensive gene flow occurring between natural olive fly populations around in Central Mediterranean basin, where Greece and Italy have been trading centers for olive products for thousands of years. The same analysis for flies from California correctly assigned 69 of the 150 individuals (46%) in their respective samples and, more importantly, 131 of the 150 (87.3%) individuals were correctly identified as Californian. Therefore, California olive flies may form a discrete subpopulation that has differentiated from the ancestral Mediterranean populations. To define the possible origin of the invasion, the five California samples were assigned to one of the three described subpopulations around the Mediterranean plus Israel. Cal-CA and Nap-CA were identified as Cypriot, whereas the remaining three were identified as Israeli (Appendix 2).
Analysis of bottleneck
Olive fly presence in California is recent (first observed in 1998), but it has expanded in the whole cultivation zone in the state. We used BOTTLENECK software to test for any recent bottleneck phenomena in the area. Results suggest that the olive fly in California has not faced any bottlenecks.
Discussion
Invasions, such as those of the olive fly, are often characterized by a unique set of demographic and genetic features that result from a small number of colonizing individuals and the rapid growth and spread of new populations (Davies and Roderick, 1999). We searched for such features in the relatively recent olive fly invasion in California to determine its origin. For this purpose, 150 wild flies captured from five different sampling areas in California were genotyped for 10 microsatellite loci. These results were integrated to those of a previous study of olive fly populations around the European part of the Mediterranean basin (Augustinos et al., 2005). In that study, three subpopulations were distinguished: Western Mediterranean (constituted of samples from the Iberian Peninsula), Central Mediterranean (samples from Greece, Italy and Turkey) and Eastern Mediterranean (samples from Cyprus).
Iberia (most likely) absolved
Normally, a colonization process goes along with a loss of genetic variability because the founder population carries only a fraction of the original population's variability. In the case of California samples, three alleles were encountered (in relatively high frequencies) that were not observed in the West Med subpopulation. Given the large number of flies analyzed from West Med, it is highly unlikely that these alleles are present in this area. In addition, there were no shared alleles between California and West Med (that is, alleles were present in these two areas but not anywhere else). Furthermore, genetic distance values are the greatest between California and West Med samples and these two areas are placed in two different nodes of a dendrogram with a bootstrap value of 100. STRUCTURE analysis also shows little possibility of California and West Med samples being in the same genetic cluster. PCA also suggests that California samples are more distantly related to the West Med ones. Finally, Bayesian assignment of groups of individuals shows that it is highly unlikely that California genotypes derive from West Mediterranean genotypes.
Greece and Italy remotely linked
The first major discrimination derived from the UPGMA dendrogram (with high bootstrap value) is the one between Eastern Med plus California samples and the rest of the Mediterranean. All analyses presented in this study show that samples from Greece, Italy and possibly Turkey form a distinct subpopulation, which is also mixed with western and/or eastern Mediterranean subpopulations, likely due to extensive gene flow in the area (that could be explained by extensive trade throughout history). STRUCTURE analysis also suggests that the Central Med cluster might be different from the cluster of Eastern Mediterranean and California, although it seems to share a certain degree of ancestry with this group. Therefore, it does not seem very likely that areas of the Central Mediterranean are the source of the California invasion. Finally, assignment of groups of individuals performed by GeneClass software gives a low probability only to the Cal-CA sample to have a Central Med origin.
East Med culpa?
Instead, all analyses of our data point to the Eastern Mediterranean as the putative source of the observed invasion. Eastern Mediterranean populations are clearly more polymorphic than California samples. Although California samples tend to form a cluster of their own, it is obvious that they are more closely related to East Med samples, either based on genetic distance values and allele frequencies or under STRUCTURE analysis and PCA. An interesting finding comes from the GeneClass analysis. When asked to assign California samples to one of the four groups assumed, three were identified as Israeli (Sol-CA, Yol-CA and SLO-CA) and the remaining two as Cypriot (Cal-CA and Nap-CA). Therefore, all were identified as East Med samples.
Olive fly in California: single or multiple invasions?
Fly collection data from the California Department of Food and Agriculture and the statewide monitoring program (through which the flies used in this analysis were obtained) suggest a rapid and virtually complete colonization by the olive fly within the range of olive cultivation in California. Till now, it remains unclear whether the fly invaded once in 1998 when it was first detected, or additional times either before the first detection or since. The fact that only a small fraction of the alleles that are found in Mediterranean are also present in California (44 out of 106) can be evidence of a flow of a small number of alleles from the Mediterranean basin toward California. Analysis of olive flies collected in subsequent years throughout the invasion time and from more collection sites will show which of the two prementioned scenarios is more likely.
References
Aketarawong N, Bonizzoni M, Thanaphum S, Gomulski LM, Gasperi G, Malacrida AR et al. (2007). Inferences on the population structure and colonization process of the invasive oriental fruit fly, Bactrocera dorsalis (Hendel). Mol Ecol 16: 3522–3532.
Augustinos AA, Mamuris Z, Stratikopoulos E, D’Amelio S, Zacharopoulou A, Mathiopoulos KD (2005). Microsatellite analysis of olive fly populations in the Mediterranean indicates a westward expansion of the species. Genetica 125: 231–241.
Augustinos AA, Stratikopoulos EE, Zacharopoulou A, Mathiopoulos KD (2002). Polymorphic microsatellite markers in the olive fly, Bactrocera oleae. Mol Ecol Notes 2: 278–280.
Bonizzoni M, Guglielmino CR, Smallridge CJ, Gomulski M, Malacrida AR, Gasperi G (2004). On the origins of medfly invasion and expansion in Australia. Mol Ecol 13: 3845–3855.
Bonizzoni M, Zheng L, Guglielmino CR, Haymer DS, Gasperi G, Gomulski LM et al. (2001). Microsatellite analysis of medfly bioinfestations in California. Mol Ecol 10: 2515–2524.
Bruford MW, Wayne RK (1993). Microsatellites and their application to population genetic studies. Cur Opin Genet Dev 3: 939–943.
California Department of Food and Agriculture (2006). California Agricultural Resource Directory. CDFA Office of Public Affairs: Sacramento, CA.
Cornuet JM, Luikart G (1997). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Davies N, Roderick GK (1999). Determining pathways of marine bioinvasion: genetic and statistical approaches. In: Pederson J (ed). First Conference of Marine Bioinvasions. Plenum Press: Boston.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Felsenstein J (1994). PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the Author. Department of Genome Sciences, University of Washington: Seattle.
Gasperi G, Bonizzoni M, Gomulski LM, Murelli V, Torti C, Malacrida AR et al. (2002). Genetic differentiation, gene flow and the origin of infestations of the medfly, Ceratitis capitata. Genetica 116: 125–135.
Gilchrist AS, Sved JA, Meats A (2004). Genetic relations between outbreaks of the Queensland fruit fly, Bactrocera tryoni (Froggatt) (Diptera: Tephritidae), in Adelaide in 2000 and 2002. Austr J Entomol 43: 157–163.
Goudet J (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available on http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995).
Katsoyannos P (1992). Olive Pests and their Control in the Near East. FAO Plant Production and Protection Paper No 115. FAO: Rome.
Mazomenos BE (1989). Dacus oleae. In: Robinson AS, Hooper G (eds). Word Crop Pests, Vol. 3B, Elsevier: Amsterdam. pp 169–177.
Meixner MD, Mcpheron BA, Silva JG, Gasparich GE, Sheppard WS (2002). The Mediterranean fruit fly in California: evidence for multiple introductions and persistent populations based on microsatellite and mitochondrial DNA variability. Mol Ecol 11: 891–899.
Nardi F, Carapelli A, Dallai R, Roderick GK, Frati F (2005). Population structure and colonization history of the olive fly, Bactrocera oleae (Diptera, Tephritidae). Mol Ecol 14: 2729–2738.
Nei M (1972). Genetic distance between populations. Am Nat 106: 283–292.
Page RDM (1996). TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358.
Peakall R, Smouse PE (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Piry S, Alapetite A, Cornuet J, Paetkau D, Baudouin L, Estoup A (2004). GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95: 536–539.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Rannala B, Mountain JL (1997). Detecting immigration by using multilocus genotypes. Proc Nat Ac Sci USA 94: 9197–9201.
Raymond M, Rousset F (1995). GENEPOP v.1.2: population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Roderick GK, Navajas M (2003). Genes in new environments: genetics and evolution in biological control. Nat Rev Gen 4: 889–899.
Rice RE (2000). Bionomics of the olive fruit fly Bactrocera (Dacus) oleae. UC Plant Protection Quarterly 10: 1–5.
Rice WR (1989). Analysis tables of statistical tests. Evolution 43: 223–225.
Schlötterer C, Pemberton J (1994). The use of microsatellites for genetic analysis of natural populations. EXS 69: 203–214.
Schneider S, Kueffer JM, Roessli D, Excoffier L (1997). ARLEQUIN, version 1.1. A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva: Switzerland.
Tautz D, Schlötterer C (1994). Simple sequences. Curr Opin Genet Dev 4: 832–837.
Yeh FC, Boyle T, Rongcai Y, Ye Z, Xiyan JM (1999). POPGENE VERSION 1.32 Microsoft Window-based Freeware for Population Genetic Analysis. http://www.ualberta.ca/~fyeh/.
Yu H, Frommer M, Robson MK, Meats AW, Shearman DCA, Sved JA (2001). Microsatellite analysis of the Queensland fruit fly Bactrocera tryoni (Diptera: Tephritidae) indicates spatial structuring: implications for population control. Bull Entomol Res 91: 139–147.
Zalom FG, Burrack HJ, Bingham R, Price R, Ferguson L (2008). Olive fruit fly (Bactrocera oleae) introduction and establishment in California. Acta Horticulturae 791: 619–627.
Acknowledgements
We thank Hannah Joy Burrack for all the California fly shipments, David Nestel for the Israeli samples and Zissis Mamuris for several useful discussions with the authors. This research was partially supported by grants to KDM from the Greek Ministry of National Education and Religious Affairs. NEZ was partially supported by a fellowship from the National Fellowship Foundation (Greece).
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1
Appendix 2
Rights and permissions
About this article
Cite this article
Zygouridis, N., Augustinos, A., Zalom, F. et al. Analysis of olive fly invasion in California based on microsatellite markers. Heredity 102, 402–412 (2009). https://doi.org/10.1038/hdy.2008.125
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.125
Keywords
This article is cited by
-
Previously introduced braconid parasitoids target recent olive fruit fly (Bactrocera oleae) invaders in Hawai’i
Scientific Reports (2023)
-
Population genetic structure of a recent insect invasion: a gall midge, Asynapta groverae (Diptera: Cecidomyiidae) in South Korea since the first outbreak in 2008
Scientific Reports (2023)
-
Exploration for olive fruit fly parasitoids across Africa reveals regional distributions and dominance of closely associated parasitoids
Scientific Reports (2021)
-
Population genetics of the soybean aphid in North America and East Asia: test for introduction between native and introduced populations
Biological Invasions (2017)
-
Genetics and biology of Anastrepha fraterculus: research supporting the use of the sterile insect technique (SIT) to control this pest in Argentina
BMC Genomic Data (2014)