Abstract
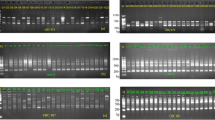
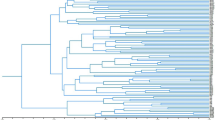
Genetic variation in chrysanthemum (Dendranthema grandiflora) was studied using a recently developed technique generating Random Amplified Polymorphic DNAs(RAPDs). It appeared that variation between cultivars was high and that the cultivars used could be distinguished from each other by using only two different primers. A family of cultivars, derived from one original cultivar by vegetative propagation, had identical fragment patterns. Because of the high level of polymorphism and clonal stability RAPD fragments are useful for cultivar identification. Genetic variability among related Dendranthema species was too high to study genetic distances either among cultivars within chrysanthemum or among species related to chrysanthemum.
Similar content being viewed by others
Article PDF
References
Aarts, J M M J G, Hontelez, J G J, Fischer, P, Verkerk, V, Van Kammen, A, and Zabel, P. 1991. Acid phosphatase-1, a tightly linked molecular marker for root-knot nematode resistance in tomato: from protein to gene, using PCR and degenerate primers containing deoxyinosine. Plant Mol Biol, 16, 657–661.
Arnold, M L, Buckner, C M, and Robinson, J J. 1991. Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc Natl Acad Sci, USA, 88, 1398–1402.
Brown, A H D. 1979. Enzyme polymorphism in plant populations. Theor Pop Biol, 15, 1–42.
Caetona-Anolles, G, Bassam, B J, and Gresshoff, P M. 1991. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Bio Technology, 9, 553–557.
Carlson, J E, Tulsieran, J K, Glaubitz, J C, Luk, V W K, Kauffeldt, C, and Rutledge, R. 1991. Segregation of random amplified DNA markers in Fl progeny of conifers. Theor Appl Genet, 83, 194–200.
Dowling, T E, Moritz, C, and Palmer, J D. 1990. Nucleic acids II: restriction site analysis. In: Hillis, D. M. and Moritz, C. (eds) Molecular Systematics, Sinauer Ass., pp. 250–317.
Dowrick, G J. 1952. The chromosomes of Chrysanthemum, I: the species. Heredity, 6, 365–375.
Dowrick, G J. 1953. The chromosomes of Chrysanthemum, II: garden varieties. Heredity, 7, 59–72.
Dowrick, G J, and El-Bayoumi, A. 1966. The origin of new forms of the garden Chrysanthemum. Euphytica, 15, 32–38.
Drewlow, L W, Ascher, P D, and Widmer, R E. 1973. Genetic studies of self incompatibility in the garden chrysanthemum, Chrysanthemum morifolium Ramat. Theor Appl Genet, 43, 1–5.
Hadrys, H, Balick, M, and Schierwater, B. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Molec Ecol, 1, 55–63.
Halward, T, Stalker, T, Larue, E, and Kochert, G. 1992. Use of single-primer DNA amplifications in genetic studies of peanut (Arachis hypogaea L.). Plant Mol Biol, 18, 315–325.
Helentjaris, T, Slocum, M, Schaefer, A, and Nienhuis, J. 1986. Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet, 72, 761–769.
Hu, J, and Quiros, C F. 1991. Identification of broccoli and cauliflower cultivars with RAPD Markers. Plant Cell Rep, 10, 505–511.
Jeffreys, A J, Wilson, V, and Thein, S L. 1985. Hypervariable ‘minisatellite’ regions in human DNA. Nature, 314, 67–73.
Kawata, J. 1978. Japanese cultivars and wild species; their useful characteristics for chrysanthemum breeding. In: Langton, F. A. (ed.) Eucarpia Meeting on Ornamentals, Littlehampton, UK, pp. 33–48.
Klein-Lankhorst, R M, Vermunt, A, Weide, R, Liharska, T, and Zabel, P. 1991. Isolation of molecular markers for tomato (L. esculentum) using random amplified polymorphic DNA(RAPD). Theor Appl Genet, 83, 108–114.
Krogulevich. 1978. In: Malyshev, L. I. and Peshlcova, G. A. (eds) Flora of the Prebaikal, Novosibirsk, pp. 19–48.
Langton, F A. 1989. Inheritance in Chrysanthemum morifolium Ramat. Heredity, 62, 419–423.
Lee, Y N. 1967. Chromosome numbers of flowering plants in Korea. J Kor Cult Res Inst, 11, 455–478.
Lee, Y N. 1975. Taxonomic study on white flowered wild chrysanthemum in Asia. J Korean Res Inst Better Living, 14, 63–79.
Loveless, M D, and Hamrick, J L. 1984. Ecological determinants of genetic structure in plant populations. Ann Rev Evol Syst, 15, 65–95.
Martin, G B, Williams, J G K, and Tanksley, S D. 1991. Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci, USA, 88, 2336–2340.
Michelmore, R W, Paran, I, and Kesseli, R V. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci, USA, 88, 9828–9832.
Nybom, H, and Hall, H K. 1991. Minisatellite DNA ‘fingerprints’ can distinguish Rubus cultivars and estimate their degree of relatedness. Euphytica, 53, 107–114.
Quiros, C F, Hu, J, This, P, Chevre, A M, and Delseny, M. 1991. Development and chromosomal localization of genome-specific markers by polymerase chain reaction in Brassica. Theor Appl Genet, 82, 627–632.
Reiter, R S, Williams, J G K, Feldmann, K A, Rafalski, J A, Tingey, S V, and Scolnik, P A. 1992. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci, USA, 89, 1477–1481.
Ronald, W G, and Ascher, P D. 1975. Self compatibility in garden Chrysanthemum: occurrence, inheritance and breeding potential. Theor Appl Genet, 46, 45–54.
Saghai-Maroof, M A, Soliman, K M, Jorgensen, R A, and Allard, R W. 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci, USA, 81, 8014–8018.
Sambrook, J, Fritsch, E F, and Maniatis, T. 1989. Molecular Cloning: a Laboratory Manual, 2nd edition. Cold Spring Harbour Laboratory Press, New York.
Shimotomai, N. 1932. Eigenartige Vermehrung der chromosomenzahl bei den Artenbastarden von Chrysanthemum. Bot Mag (Tokyo), 46, 789–799.
Stephens, L C, Ascher, P D, and Widmer, R E. 1984. Interaction among sporophytic S loci in self-incompatible garden Chrysanthemums. Euphytica, 33, 623–631.
Swofford, D L, and Olsen, G J. 1990. Phylogeny reconstruction. In: Hillis, D. M. and Moritz, C. (eds) Molecular Systematics, Sinauer Ass. pp. 411–501.
Teynor, T M, Ascher, P D, Widmer, R E, and Luby, J J. 1989. Inheritance of flower color in Dendranthema grandiflora Tzvelev. (Chrysanthemum morifolium Ramat.) using cultivars and inbreds II. Vacuole pigmentation. Euphytica, 42, 297–305.
Van Heusden, A W, and Bachmann, K. 1992. Genotype relationships in Microseris elegans (Asteraceae, Lactuceae) revealed by DNA amplification from arbitrary primers (RAPDs). Pl Syst Evol, 179, 221–233.
Watanabe, K. 1977. The control of diploid-like meiosis in polyploid taxa of Chrysanthemum (Compositae). Jap J Genetics, 52, 125–131.
Weining, S, and Langridge, P. 1991. Identification and mapping of polymorphisms in cereals based on the polymerase chain reaction. Theor Appl Genet, 82, 209–216.
Welsh, J, and McClelland, M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res, 18, 7213–7218.
Welsh, J, Honeycutt, R J, McClelland, M, and Sobral, B W S. 1991. Parentage determination in maize hybrids using the arbitrarily primed polymerase chain reaction (AP-PCR). Theor Appl Genet, 82, 473–476.
Williams, J G K, Kubelik, A R, Livak, K J, Rafalski, J A, and Tingey, S V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res, 18, 6531–6535.
Wolff, K. 1991. Analysis of allozyme variability in three Plantago species and a comparison to morphological variability. Theor Appl Genet, 81, 119–126.
Wolff, K, Schoen, E D, and Peters-Van Rijn, J. 1993. Optimizing the generation of Random Amplified Polymorphic DNAs in chrysanthemum (Dendranthema grandiflora Tzvelev.). Theor Appl Genet, in press.
Wu, K K, Burnquist, W, Sorrells, M E, Tew, T L, Moore, P H, and Tanksley, S D. 1992. The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theor Appl Genet, 83, 294–300.
Zagorski, J S, Ascher, P D, and Widmer, R E. 1983. Multigenic self incompatibility in hexaploid Chrysanthemum. Euphytica, 32, 1–7.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Wolff, K., Peters-van Rijn, J. Rapid detection of genetic variability in chrysanthemum (Dendranthema grandiflora Tzvelev) using random primers. Heredity 71, 335–341 (1993). https://doi.org/10.1038/hdy.1993.147
Received:
Issue Date:
DOI: https://doi.org/10.1038/hdy.1993.147