Abstract
Loss of Ras association domain family protein 1 isoform A (RASSF1A) expression is associated with the development of a variety of human cancers and the expression of carcinoembryonic antigen (CEA) frequently occurs in gastric cancer. This study investigated the effects of RASSF1A expression restoration using a hypoxia-inducible CEA promoter-driven vector on xenograft tumor growth in nude mice and on the in-vitro regulation of gastric cancer cell viability, cell cycle distribution, apoptosis, colony formation and invasion capacity. The data showed that the level of CEA mRNA and protein was much higher in gastric cancer SGC7901 cells than in a second gastric cancer cell line, MKN28, or in the MCF-10A normal epithelial breast cell line. RASSF1A expression was restored in SGC7901 cells compared with the negative control virus-infected SGC7910 cells. RASSF1A expression restoration significantly inhibited gastric cancer cell viability, colony formation and invasion capacity, but induced cell cycle arrest and apoptosis in vitro, especially under hypoxic culture conditions. At the gene level, restoration of RASSF1A expression under hypoxic culture conditions significantly suppressed matrix metalloproteinase-2 expression and prevented cyclinD1 expression. A nude mouse xenograft assay showed that the restoration of RASSF1A expression reduced gastric cancer xenograft formation and growth. In conclusion, the restoration of RASSF1A expression using a hypoxia-inducible and CEA promoter-driven vector suppressed aggressive phenotypes of gastric cancer cells in vitro and in vivo. These results suggest that LV-5HRE-CEAp-RASSF1A gene therapy may be a promising novel approach to treat advanced gastric cancer.
Similar content being viewed by others
Introduction
Gastric cancer is one of the most commonly diagnosed cancers and is the most frequent cause of cancer death in the world.1, 2 Although there have recently been significant advancements in gastric cancer treatment, including new surgical, chemotherapeutic and radiotherapeutic methods, many patients are still dying after being diagnosed with advanced gastric cancer and developing metastasized disease.3 Thus, gastric cancer remains a major clinical challenge. Tumor-targeted gene therapy may be a promising new approach for the effective control of gastric cancer. Gene therapy is defined as the use of vectors to bring nucleic acids into cells to alter gene expression and prevent, halt or reverse a pathological process. The number of ongoing or completed gene therapy clinical trials approved worldwide since 1996 has reached 1274 and 63.8% of these were for human cancer.4 Although significant progress has been made in the development and use of tumor-selective delivery systems in the past 20 years, there are still many challenges that must be overcome to develop therapeutic genes that are specifically expressed in cancer cells, but not in normal cells. To achieve this goal, strategies have been developed that include the use of specific gene promoters, enhancers and 5′-untranslated region that respond to tumor-specific transcription factors, to drive or knock down gene expression to treat human cancers.
We focused on the Ras association domain family protein 1 isoform A (RASSF1A), because lost RASSF1A expression is associated with the development of a variety of human cancers, including gastric cancer or other gastrointestinal malignancies, and this suggests the possibility that RASSF1A may have a tumor suppressor function in these human cells.5, 6, 7 Restoration of RASSF1A expression, therefore, could potentially control gastric cancer progression. Moreover, carcinoembryonic antigen (CEA) is a glycosylated protein that is also highly expressed in a number of cancers, including gastric cancer.8 The CEA promoter has been used in many cancer studies and can specifically target CEA-positive cancer cells.9, 10 Hypoxia-inducible factors (HIFs) are another potential target. Hypoxia frequently occurs in solid tumor lesions.11 Accumulating data shows that 50%–60% of locally advanced solid tumors develop hypoxia.12 Hypoxia promotes tumor progression, invasion of adjacent tissues and metastasis, and hypoxic tumors are resistant to radiotherapy and chemotherapy.13 HIF-1, a well-studied HIF, is a heterodimer of the HIF-1α and HIF-1β subunits. One subunit, HIF-1α, is an O2-regulated subunit that induces HIF-1 activity and expression in response to a reduction of O2 availability, whereas the other, HIF-1β, is a constitutive subunit.14 HIF-1α binds to the hypoxia-responsive element (HRE), a conserved transcription promoting sequence in the regulatory regions of target genes.15 Previous studies have shown that HIF-1α contributes to the malignant phenotypes of gastric cancer.16, 17, 18
In this study, to restore RASSF1A expression, we constructed an expression vector carrying RASSF1A under the control of hypoxia-inducible and CEA promoters, to specifically restore RASSF1A expression in gastric cancer cells. We then assessed the effects of RASSF1A expression on xenograft tumor growth in nude mice and the regulation of gastric cancer cell viability, cell cycle distribution, apoptosis, colony formation and invasion capacity in vitro. The results of this study could help the future development of an inducible tumor-specific lentiviral vector that combines RASSF1A with CEAp and 5HRE (LV-5HRE-CEAp-RASSF1A) for gastric cancer gene therapy.
Results
Expression of endogenous CEA and RASSF1A mRNA and protein in gastric cancer cells
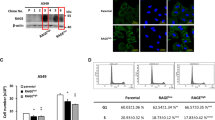
We first detected expression of endogenous CEA mRNA using quantitative reverse-transcription PCR in several gastric cancer cell lines as well as in a normal breast cell line. The results showed that the level of CEA mRNA in gastric cancer SGC7901 cells was much higher than that in both the MKN28 gastric cancer and normal breast MCF-10A cell lines (Figure 1a). The same was true for endogenous CEA protein (Figure 1b). The mean density of the CEA protein, as quantified using the immunohistochemical staining results, was 0.111±0.024, 0.076±0.016 and 0.002±0.000 in SGC7901, MKN28 and MCF-10A cells, respectively (P<0.001 between SGC7901 or MKN28 and MCF-10A cells; P<0.01 between SGC7901 and MKN28 cells). These data clearly indicate that the level of endogenous CEA protein was highest in SGC7901 cells, moderate in MKN28 cells and lowest in MCF-10A cells. Furthermore, endogenous RASSF1A protein was not expressed in SGC7901 or MKN28 cells, but was expressed in MCF-10A cells (Figure 1c). Thus, due to the levels of CEA and RASSF1A expression, we selected SGC7901 cells (CEA mRNA+, CEA protein+, RASSF1A protein−) for the experimental group and MKN28 cells (CEA mRNA−, CEA protein+, RASSF1A protein−) for the negative control.
Expression of endogenous CEA and RASSF1A mRNA or protein in SGC7901, MKN28 and MCF-10A cells. (a) Quantitative reverse-transcription (qRT-PCR) analysis of CEA mRNA levels ***P<0.001 vs MKN28 and MCF-10A. (b) Quantified data for CEA protein levels in SGC7901, MKN28 and MCF-10A cells. (c) Western blot analysis of RASSF1A protein levels in SGC7901, MKN28 and MCF-10A cells. ***P<0.001 vs MCF-10A; ##P<0.01 vs MKN28.
Restoration of RASSF1A expression in gastric cancer cells using a CEAp or 5HRE-driven expression vector
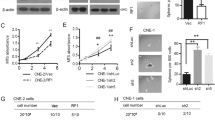
We stably restored RASSF1A expression in gastric cancer cells using CEAp or 5HRE-driven expression vectors. RASSF1A expression in SGC7901/5HC-RASSF1A and MKN28/5HC-RASSF1A cells was confirmed by quantitative reverse-transcription PCR and western blotting (Figures 2a and b). SGC7901/5HC-RASSF1A cells showed very high levels of RASSF1A mRNA under hypoxic conditions (300 μm l−1 cobalt chloride (CoCl2)), but only slightly elevated levels of RASSF1A mRNA under normoxic conditions. Similarly, RASSF1A protein levels were significantly increased under hypoxic conditions. However, RASSF1A expression was low in SGC7901, SGC7901/NC, MKN28/5HC-RASSF1A and MKN28 cells under both normoxic and hypoxic conditions. These data indicated that both RASSF1A mRNA and protein were expressed in SGC7901/5HC-RASSF1A cells, especially under hypoxic conditions.
Overexpression of RASSF1A mRNA in SGC7901 and MKN28 cells. (a) Quantitative reverse transcription (qRT-PCR) analysis of RASSF1A mRNA in RASSF1A stably transfected cells. ***P<0.001 vs SGC7901, SGC7901/NC, MKN28 and MKN28/5HC-RASSF1A cells; •••P<0.001 vs SGC7901/5HC-RASSF1A cells cultured under normoxic conditions. (b) Western blot analysis of RASSF1A protein in RASSF1A stably transfected cells under normoxic (N) and hypoxic (H) conditions. (c) Luciferase activity assay. 5HRE-CEAp activity was measured in SGC7901, MKN28 and MCF-10A cells. *P<0.05 vs MKN28 and MCF-10A cells under hypoxic conditions; **P<0.001 vs MKN28 cells under normoxic conditions; ***P<0.001 vs SGC7901 under normoxic conditions.
The hypoxic inducibility, CEA-targeting and RASSF1A expression-driving ability of the 5HRE-CEAp element was confirmed using a dual luciferase assay (Figure 2c). SGC7901, MKN28 and MCF-10A cells were transfected with pGL4.20-5HRE-CEAp-Luc or control vectors and cultured for 24 h under normoxic or hypoxic conditions. The luciferase activity fold change was measured and the data showed a significant increase in luciferase activity in SGC7901 cells under normoxic conditions (P<0.05; 59.63±24.24 in SGC7901 vs 27.82±7.19 in MKN28 and 1.25±0.57 in MCF-10A). However, luciferase activity in SGC7901 and MKN28 cells was further increased in the presence of CoCl2 (300 μm l−1) when compared with luciferase activity under aerobic conditions (SGC7901, P<0.001; 519.75±97.14 vs 59.63±24.24; MKN28 P<0.001; 87.80±7.78 vs 27.82±7.19).
Effects of RASSF1A expression on regulation of gastric cancer cell viability, apoptosis, cell cycle distribution, colony formation and invasion capacity in vitro
The viability of cells expressing RASSF1A was assessed using a CCK-8 assay. The data showed that RASSF1A expression significantly reduced SGC7901/5HC-RASSF1A cell viability when compared with either SGC7901 or SGC7901/NC cell viability under both normoxic and hypoxic conditions. Under hypoxic conditions, RASSF1A expression further decreased tumor cell viability (Figures 3a and b). However, this result was not replicated in MKN28/5HC-RASSF1A cells (Figure 3b), because MKN28 cells do not express CEA and, thus, the vector did not restore RASSF1A expression in these cells (Figure 2b).
Effects of RASSF1A expression on SGC7901 and MKN28 cell viability. (a) Cell viability CCK-8 assay. SGC7901, SGC7901/NC and SGC7901/5HC-RASSF1A cells were grown in monolayer with or without CoCL2 for 5 days and then subjected to cell viability analysis. (b) Growth inhibition of SGC7901, SGC7901/5HC-RASSF1A, MKN28 and MKN28/5HC-RASSF1A cells with or without CoCL2 on the fourth day after treatment. **P<0.01 vs SGC7901/5HC-RASSF1A and SGC7901-CoCL2; *P>0.05 vs MKN28/5HC-RASSF1A.
Moreover, a tumor cell apoptosis assay showed that the rate of apoptosis was increased in SGC7901/5HC-RASSF1A cells under hypoxic conditions (Figure 4a). Cell cycle analysis revealed that SGC7901/5HC- RASSF1A cells accumulated at the G1 phase of the cell cycle under hypoxic, but not normoxic, conditions (P<0.05; Figure 4b). Colony formation assays also showed a significant reduction in the numbers of colonies that were formed on the plate by SGC7901/5HC-RASSF1A cells under hypoxic conditions (P<0.05; Figures 5a and b). In addition, the Transwell assay showed that the number of tumor cells passing through the Matrigel-coated membrane was significantly reduced in the SGC7901/5HC-RASSF1A cells (P<0.01) and was further reduced under hypoxic conditions (Figures 5c and d), suggesting that RASSF1A expression suppressed gastric cancer cell invasion.
Effect of RASSF1A expression on the regulation of SGC7901 cell apoptosis and cell cycle distribution under normoxic or hypoxic conditions. (a) Flow cytometry apoptosis assay. SGC7901 cells were grown and subjected to tumor cell apoptosis analysis. **P<0.01 vs SGC7901 and SGC7901/NC cells under normoxic or hypoxic conditions and SGC7901/5HC-RASSF1A cells under normoxic conditions. (b) Flow cytometric cell cycle analysis. Cells were grown under normoxic or hypoxic conditions and subjected to cell cycle analysis.
Effect of RASSF1A expression on the regulation of tumor cell colony formation and migration capacity in normoxic (N) or hypoxic (H) conditions. (a) Colony formation assay. SGC7901, SGC7901/NC and SGC7901/5HC-RASSF1A cells were grown under normoxic or hypoxic conditions and subjected to colony formation assay. (b) Summarized data of a. *P<0.05 vs SGC7901 or SGC7901/NC cells under hypoxic conditions. (c) Transwell tumor cell migration assay. SGC7901, SGC7901/NC and SGC7901/5HC-RASSF1A cells were grown under normoxic or hypoxic conditions and subjected to a tumor cell migration assay. (d) Summarized data of c. ★★P<0.01 vs SGC7901 or SGC7901/NC cells under normoxic conditions; **P<0.01 vs SGC7901/5HC-RASSF1A cells under normoxic conditions.
Effects of RASSF1A expression on the regulation of the HIF-1α, caspase 3, MMP-2 and cyclinD1 proteins in gastric cancer cells
To investigate the molecular events associated with recombinant lentivirus vector pLV-5HRE-CEAp-RASSF1A-mediated RASSF1A expression in gastric cancer cells, we assessed whether restoring RASSF1A expression under hypoxic conditions could affect the expression of HIF-1α, caspase 3, matrix metalloproteinase-2 (MMP-2) and cyclinD1 proteins in SGC7901 cells. HIF-1α protein was not detected in SGC7901, SGC7901/NC or SGC7901/5HC-RASSF1A cells under normoxic conditions; however, expression could be equally detected in all cell lines under hypoxic conditions (Figure 6a). Conversely, MMP-2 protein levels were significantly reduced under hypoxic conditions (P<0.01; Figures 6a and b). Although caspase 3 protein showed no expression differences in SGC7901, SGC7901/NC or SGC7901/5HC-RASSF1A cells under normoxic conditions, its expression only increased under hypoxic conditions in SGC7901/5HC-RASSF1A cells (P<0.01; Figures 6a–c). CyclinD1 expression was increased under hypoxic conditions when compared with normoxic conditions, but RASSF1A expression prevented cyclinD1 expression under hypoxic conditions (P<0.01; Figures 6a–d).
Effect of RASSF1A expression on the regulation of HIF-1α, caspase 3, MMP-2 and cyclinD1 protein expression in SGC7901 cells under normoxic (N) or hypoxic (H) conditions. (a) Western blot analysis of HIF-1α, caspase 3, MMP-2, cyclinD1 and β-actin in SGC7901, SGC7901/NC and SGC7901/5HC-RASSF1A cells. β-Actin was used as the internal control. (b) Summarized data of MMP-2 protein in cells. **P<0.01 vs SGC7901 or SGC7901/NC cells under hypoxic conditions and SGC7901/5HC-RASSF1A cells under normoxic conditions; (c) Summarized data of caspase 3 protein in cells. **P<0.01 vs SGC7901 or SGC7901/NC cells under hypoxic conditions; (d) Quantification of cyclinD1 protein expression in cells, **P<0.01 vs SGC7901/5HC-RASSF1A cells under normoxic conditions.
Effect of RASSF1A on gastric cancer xenograft formation and growth in nude mice
SGC7901 cells were subcutaneously implanted in BALB/c nude mice. Xenograft tumors formed within 8 days and grew to 50–100 mm3. We injected lentiviruses (1 × 108 pfu) in 100 μl of serum-free medium intratumorally at three sites per xenograft tumor and repeated the injections on day 4. Two weeks after the second virus injection, mice were killed and xenograft tumors were collected, measured and photographed. Compared with the negative solution and LV-NC (pLV-5HRE-CEAp-NC) groups, the size and weight of the tumors were significantly lower in the LV-5HC-RASSF1A-treated mice (P<0.01; Figure 7).
Effect of RASSF1A expression on the regulation of tumorigenicity in nude mice. (a) The mean tumor volume at the end of experiments. **P<0.01 vs NS and LV-NC groups. (b) The mean tumor weight at the end of experiments. **P<0.01 vs NS and LV-NC groups. (c) Image of xenograft tumors from each group. (d) Immunohistochemical analysis of RASSF1A expression in xenograft tumors and normal mouse liver tissues.
Immunohistochemical data also confirmed RASSF1A expression in gastric cancer cell xenografts. As shown in Figure 7d, RASSF1A expression was lower in the negative solution-injected or LV-NC-injected xenograft tumors, but RASSF1A expression was restored in xenograft tumors after the LV-5HRE-CEAp-RASSF1A injection. It is also true that there was no RASSF1A expression in normal mouse hepatocytes in the liver tissues (Figure 7d).
Discussion
Gene therapy is a promising option for the treatment human cancers; however, two keys to success, a tumor-selective delivery system and the persistent expression of anti-cancer gene products, remain elusive. In the current study, we developed a lentivirus vector for hypoxia-inducible, CEA promoter-driven and tissue-specific expression of RASSF1A, LV-5HRE-CEAp-RASSF1A. Assessing this vector showed that the following: (i) the regulatory element 5HRE-CEAp can induce luciferase reporter activity under hypoxic conditions in gastric cancer SGC7901 cells expressing high levels of CEA; (ii) RASSF1A expression levels were induced in RASSF1A-negative gastric cancer SGC7901 cells under hypoxic conditions; (iii) the LV-5HRE-CEAp-RASSF1A lentivirus suppressed aggressive SGC7901 gastric cancer cell phenotypes under hypoxic conditions, including cell viability, apoptosis, cell cycle arrest, colony formation and invasion capacity; (iv) RASSF1A expression regulated the expression of the caspase 3, MMP-2 and cyclinD1 proteins; and (v) LV-5HRE-CEAp-RASSF1A infection suppressed gastric cancer xenograft tumor formation and growth in nude mice. Thus, our current study provided a gene (RASSF1A is frequently lost in gastric cancer or other gastrointestinal cancers) and specific promoter-driven RASSF1A expression (CEA is usually overexpressed in these cancers) for future control of gastric cancer. Thus, our current data strongly supports the potential usefulness of LV-5HRE- CEAp-RASSF1A as an effective anti-tumor treatment option for advanced gastric cancer.
A tumor-selective delivery system is the key to successful tumor gene therapy. Over time, many gene therapy strategies have been developed and these strategies have been accompanied and shaped by an increasing knowledge of tumor pathogenesis. Hypoxia, caused by rapid tumor growth, has a key role in cancer progression and is the focus of many cancer treatment strategies.19 For example, Shibata et al.20 constructed an NTR/CB1954 vector under the control of five copies HRE and a minimal cytomegalovirus promoter (CMV) promoter (5HRE/CMVmp) and confirmed specific anti-tumor activity under hypoxic conditions. They showed that among various HRE constructs and promoter elements, 5HRE/CMVmp had optimal activation at a low oxygen tension, which is true in the gastrointestinal tract. Harvey et al.21 developed an adenovirus vector for hypoxia-targeted gene therapy using the herpes simplex virus thymidine kinase and bacterial nitroreductase prodrug-activating genes. They showed that 5HRE derived from vascular endothelial growth factor and linked to the CMV minimal promoter could induce optimum luciferase reporter gene expression.
In the current study, we constructed a lentiviral vector under the control of a 5HRE element and a CEA promoter to enhance RASSF1A expression in tumor lesions. This 5HRE regulatory element showed hypoxia-inducible activity and its design conformed to the HRE described by Shibata et al.20 In addition to a hypoxia-driven strategy for tumor targeting, the inclusion of the CEA promoter serves as a secondary tumor targeting strategy. The CEA promoter has been used in many studies, is highly expressed in a number of tumors and is a known tumor marker of digestive cancer. Nyati et al.22 used the CEA promoter and enhancer to selectively drive prodrug yeast cytosine deaminase expression in CEA-positive cancer cells and demonstrated that their vector had the ability to suppress tumor growth. Liu et al.23 constructed a CMV enhancer and CEA promoter regulatory element, and demonstrated the ability to specifically target CEA-positive gastric cancer. Although the CEA promoter was used as part of a strategy of specific cancer gene delivery, the CEA promoter alone could only weakly drive target transgene expression, leading to insufficient anti-cancer gene expression for cancer gene therapy.24 To overcome this challenge, various enhancers were added in front of the CEA promoter.22, 23, 24 We used 5HRE as the CEA promoter enhancer.
Although it has only been studied for 14 years, RASSF1A is reported to be a putative tumor suppressor gene. Dammann et al.25 showed that RASSF1A is localized at chromosome 3p21.3 and that it was inactivated by methylation of the CpG-island promoter. They also showed that RASSF1A expression was lost in lung tumors and re-expression of RASSF1A had anti-oncogenic effects in lung carcinoma cells. Since then, accumulating data has demonstrated that through multi-mechanisms, RASSF1A is a tumor suppressor gene in the vast majority of human cancers.26 Our data, reported here, also demonstrated that RASSF1A has effective anti-tumor activity. Shivakumar et al.27 showed that RASSF1A expression blocked cell cycle progression at the G1/S phase by inhibiting cyclinD1 accumulation. Agathanggelou et al.28 found that transfection of RASSF1A cDNA into A549 cells induced cell cycle arrest at the G1 phase and downregulated cyclinD1 expression. Our current study further confirmed these results and showed that the effects were significantly increased under hypoxic culture conditions. However, the cell cycle impact of RASSF1A under hypoxic conditions may be due to other unknown mechanisms. Apoptosis promotion is an important tumor suppressor function of RASSF1A and many studies have investigated the pro-apoptotic mechanisms of RASSF1A.29, 30, 31, 32 Our current study demonstrated the effect of RASSF1A on the induction of SGC7901 apoptosis under hypoxic conditions. Furthermore, other studies have demonstrated that RASSF1A can suppress the migration of tumor cells.33 Our current study revealed that the mechanism by which RASSF1A inhibits the invasive capacity of SGC7901 cells under hypoxic conditions might be related to the inhibition of MMP-2 expression. Although hypoxia can induce tumor progression, our current data indicate that LV-5HRE-CEAp-RASSF1A can effectively use that same hypoxia to drive the anti-tumor activity of RASSF1A.
Our current study is just ‘proof-of-principle’ and much more work must be done before our results can be translated into clinical trials, such as safety and efficiency of this lentivirus. Our LV-5HRE-CEAp-RASSF1A system may help to solve two key gene therapy problems in cancer, specificity and efficiency. We showed that this system can induce gene expression in CEA-positive gastric cancer cells under hypoxic conditions, further ensuring the cancer cell-specific expression of therapeutic genes. In addition, this study represents the first successful use of the tumor suppressor gene RASSF1A in gene therapy and confirms that the expression of RASSF1A effectively inhibits the growth of gastric cancer cells in vivo and in vitro. More work should be done in the future to verify the persistence and specificity of RASSF1A gene expression using the system described in this study.
Materials and methods
Cell lines and cultures
All cell lines were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Human gastric adenocarcinoma cell lines (SGC-7901 and MKN28) were grown in RPMI1640 (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal calf serum, 100 units per ml penicillin and 100 units per ml streptomycin in a humidified incubator with 5% CO2 at 37 °C. Human normal mammary epithelial cell line MCF-10A was maintained in Dulbecco’s modified Eagle’s medium/F12 (1:1) medium (Life Technologies) supplemented with 15% fetal calf serum, 10 μg ml−1 insulin, 20 ng ml−1 epidermal growth factor, 100 units per ml penicillin and 100 units per ml streptomycin in a humidified incubator with 5% CO2 at 37 °C. For hypoxic cell culture, cells were incubated with medium containing a hypoxia-mimicking agent, CoCl2.
Amplification and generation of target DNA fragments
RASSF1A cDNA fragment (NM_007182), 5HRE and CEAp were amplified from the pcDNA-RASSF1A (GeneChem Co., Ltd, Shanghai, China) and pLEGFP-N1-5HRE-CEAp-TSST-1-linker-CD80TM,34 respectively, using PCR with the specific primers listed in Table 1. All primers were synthesized by Sangon Biotech (Shanghai, China). The 5HRE-CEAp products were digested with BglII/HindIII, whereas the RASSF1A products were digested with HindIII/KpnI. The 5HRE-CEAp-RASSF1A (5HC- RASSF1A) fragment was ligated using T4 DNA ligase to link the 5HRE-CEAp and RASSF1A fragments containing HindIII/KpnI sites. However, negative control vector just contains all 5HRE-CEAp promoter sequences without RASSF1A cDNA. All sequences were verified by DNA sequence analysis (BGI·Tech, Shenzhen, China).
Lentivirus infection of SGC7901 and MKN28 cells
A lentiviral vector carrying RASSF1A, pLV-5HRE-CEAp-RASSF1A or a negative control vector, pLV-5HRE-CEAp-NC, and corresponding viruses (1 × 108 pfu) were custom constructed and prepared by GeneChem Co., Ltd. Lentivirus infection was performed in the presence of polybrene (GeneChem Co., Ltd.) according to the manufacturer’s protocol. Specifically, cells were seeded in 24-well plates overnight at a density of 5 × 104 cells per well and then infected with pLV-5HRE-CEAp-RASSF1A or pLV-5HRE-CEAp-NC lentivirus at a multiplicity of infection of 10 pfu per cell. Three days later, cells were further cultured in a medium containing 2 μg ml−1 puromycin for SGC7901 and 10 μg ml−1 for MKN28 for 2 weeks. The drug-resistant clones were used in the following experiments.
Quantitative reverse-transcription PCR
Stably transfected SGC7901/5HC-RASSF1A, SGC7901/NC or MKN28/5HC-RASSF1A cells were subjected to total cellular RNA isolation using an RNA fast 200 kit (Pioneer Biotech, Xi’an, China) according to the manufacturer’s instructions. The resulting RNA was reverse transcribed into cDNA using PrimeScript RT Master Mix (Takara, Dalian, China), according to the manufacturer’s instructions, and then subjected to quantitative reverse-transcription PCR analysis of RASSF1A mRNA using SYBR Premix Ex Taq II (Takara) and specific primers (Table 1). Each reaction was performed in triplicate and the mean RASSF1A mRNA level in each cell line was calculated by the 2−ΔΔCt method.
Immunocytochemistry
Cells were cultured in 24-well plates with 1 × 104 cells per well for 48 h and then washed with phosphate buffered saline and fixed with 4% paraformaldehyde for 30 min. CEA expression was detected by using a CEA polyclonal antibody (1:100, Proteintech, Chicago, IL, USA) and SP assay and DAB detection kits (ZSGB-BIO, Beijing, China). The negative control used an equal volume of phosphate buffered saline to replace the primary antibody. CEA immunostaining quantification was performed using digital image analysis with the Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA) according to the methods described in a previous study.35 Briefly, images were acquired by a Nikon ECLIPSE Ti-S microscope mounted with a Nikon digital camera (Nikon, Tokyo, Japan). Five randomly selected fields (× 400) were acquired per well. The integrated optical density of the positive field and area of interest was measured. The mean density (optical density/area of interest) represented the quantification of the specific protein per unit area.
Protein extraction and western blotting
Cells were grown and treated with or without CoCl2 before homogenization in RIPA buffer containing a protease inhibitor cocktail (Roche, Mannheim, Germany). Cell lysates containing 30 μg total protein were electrophoresed on a 10% SDS–polyacrylamide gel and transferred to polyvinylidene difluoride membranes. The membranes were blotted with anti-HIF-1α, caspase 3, MMP-2, cyclinD1, β-actin monoclonal antibody (Epitomics, Burlingame, USA) or an anti-RASSF1A monoclonal primary antibody (eBioscience, San Diego, USA) at a dilution of 1:1000, followed by goat anti-Rabbit horseradish peroxidase secondary antibody (ABGENT, San Diego, USA). β-Actin was probed with an anti-β-actin monoclonal antibody conjugated with horseradish peroxidase (San Ying Biotechnology, Wuhan, China). Protein bands were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Biotechnology, Rockford, IL, USA).
Luciferase activity assay
To assess pGL4.20-5HRE-CEAp-Luc activity under both normoxic and hypoxic conditions, we constructed reporter vectors based on the vector pGL4.74 (hRluc/TK) and used the Dual-Glo Luciferase Assay System E2920 (Promega, Madison, WI, USA). The promoterless firefly luciferase reporter gene vector pGL4.20 was used as a negative control. The pGL4.74 (hRluc/TK) vector containing the hRluc (Renilla reniformis) luciferase reporter gene and a herpes simplex virus thymidine kinase promoter were used as internal controls. 5HRE-CEAp was inserted into multiple cloning sites of the pGL4.20 vector, resulting in the pGL4.20-5HRE-CEAp-Luc recombinant plasmid. For the luciferase activity assay, log-phase growing cells were cultured in 96-well plates with 5 × 103 cells per well. When the cells reached the optimal 70%–90% confluency, the cells were transfected with 200 ng of pGL4.20 or pGL4.20-5HRE-CEAp-Luc plus pGL4.74 (30 ng) using TurboFect Transfection Reagent (Thermo Scientific, Vilnius, Lithuania) according to the manufacturer’s protocols. Twenty-four hours after transfection, cells were treated with or without the CoCl2 hypoxia-mimicking agent for an additional 24 h and collected for the dual luciferase assay using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocols. Luminescence was measured using a PerkinElmer EnSpire Multilabel Reader 2300 (PerkinElmer Inc., Waltham, MA, USA). Firefly luciferase activity was normalized to Renilla luciferase activity. For easy comparison, fold changes were used and the data were summarized using the following formulation: sample (Firefly luciferase activity/Renilla luciferase activity)/control (Firefly luciferase activity/Renilla luciferase activity).
Cell viability assay
A cell counting kit (CCK-8, Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assay cell viability. Briefly, 2000 viable and stably gene-transfected cells per well were seeded into 96-well tissue culture plates to a final volume of 200 μl of the growth medium. The hypoxia group was treated with CoCl2 after 24 h. The concentration of CoCl2 was optimized by CCK-8 assay, for example, 50 μm l−1 for SGC7901 and 100 μm l−1 for MKN28. After treatment and incubation, each plate was subjected to the CCK-8 assay according to manufacturer’s protocols. After 2 h incubation at 37 °C, absorbance at 450 nm was measured with PerkinElmer EnSpire Multilabel Reader 2300 (PerkinElmer Inc.). The data were plotted in a graph (Figure 3) and summarized as the percent of growth inhibition.
Flow cytometric apoptosis and cell cycle distribution assays
SGC7901, SGC7901/5HRE-CEAp-RASSF1A and SGC7901/NC cells were seeded into 60-mm culture dishes and the hypoxia group was treated with CoCl2 (300 μm l−1) for 12 h. For apoptotic analysis, a FITC Annexin V Apoptosis Detection Kit I (Becton Dickinson, Franklin Lakes, NJ, USA) was used to analyze apoptosis according to the manufacturer’s instructions. For cell cycle analysis, duplicate cells were fixed overnight with 75% ethanol at −20 °C, incubated with RNase A at 37 °C for 30 min and then incubated with propidium iodide at room temperature for 30 min. Cells were examined by flow cytometry and the data were analyzed by CellQuest version 3.3 software (Becton Dickinson).
Colony formation assay
SGC7901, SGC7901/5HRE-CEAp-RASSF1A and SGC7901/NC cells were seeded in 6-well plates at a density of 200 cells per well. All wells were cultured for 2 weeks in selection medium containing 1 μg ml−1 puromycin. The hypoxic group was also treated with CoCl2 (50 μm ml−1) and, after treatment and incubation, cells were fixed and stained with 1% crystal violet and the cell colonies were counted under an inverted microscope.
Tumor cell invasion assay
A Transwell system was used to assess tumor cell invasion capacity. In brief, upper chamber filters were coated with 50 μl BD Matrigel (Becton Dickinson) according to the manufacturer’s instructions; serum-starved cells were trypsinized and plated at 5 × 104 cells per upper chamber of a 24-well plate (8 μm, Corning, New York, NY, USA) and then the lower chambers were filled with 10% fetal bovine serum. For the hypoxia group, CoCl2 (300 μm l−1) was also added to the upper chamber. The cells were allowed to incubate 30 h at 37 °C. After incubation, the non-invasive cells were removed from the upper chamber surface with a cotton swab and the cells under the lower surface were fixed with 4% paraformaldehyde for 20 min and stained with 1% crystal violet solution. Cell invasion was quantified by counting the number of cells in five fields per well under a microscopy.
Nude mouse xenograft assay
SGC7901 cells were subcutaneously injected into the right flank of 6-week-old BALB/c nude mice at a density of 3 × 106 in 100 μl phosphate buffered saline. Tumor volume was calculated using the empirical formula V=1/2 × ((the shortest diameter)2 × (the longest diameter)). After xenograft tumors had reached 50–100 mm3, mice were randomly divided into three groups: negative solution, LV-NC and LV-5HRE-CEAp-RASSF1A (LV-5HC-RASSF1A). Lentiviruses (1 × 108 pfu) in 100 μl serum-free medium were administered intratumorally at three sites per tumor. Three days later, lentivirus administration was repeated. In the negative solution group, 100 μl serum-free RPMI1640 was injected twice instead of the recombinant virus. Tumor size and growth were monitored and measured using a caliper at regular intervals. Mice were killed 2 weeks after the second virus injection and xenograft tumors were collected, measured and photographed. Xenograft tumors were fixed in 10% buffered formalin and embedded in paraffin for sectioning and immunohistochemical staining of RASSF1A expression.
Statistical analysis
All data are representative of three independent experiments and are presented as the mean±s.d. A two-sample t-test was performed to analyze two independent samples, whereas analysis of variance was conducted for comparison among groups. SPSS 13.0 software (SPSS, Chicago, IL, USA) was used to calculate the P-value and a P<0.05 was considered to be statistically significant.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Penon D, Cito L, Giordano A . Novel findings about management of gastric cancer: a summary from 10th IGCC. World J Gastroenterol 2014; 21: 8986–8992.
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ . Gastric cancer. Lancet 2009; 374: 477–490.
Gene Therapy Clinical Trials Worldwide, Gene Med. Available at http://www.wiley.com//legacy/wileychi/genmed/clinical (updated January 2014).
Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A et al. The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A -induced cell death. J Biol Chem 2004; 279: 29247–29254.
Liu L, Vo A, McKeehan WL . Specificity of the methylation-suppressed A isoform of candidate tumor suppressor RASSF1 for microtubule hyperstabilization is determined by cell death inducer C19ORF5. Cancer Res 2005; 65: 1830–1838.
Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ . Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem 2000; 275: 35669–35672.
Hammarström S . The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol 1999; 9: 67–81.
Li Y, Chen Y, Dilley J, Arroyo T, Ko D, Working P et al. Carcinoembryonic antigen-producing cell-specific oncolytic adenovirus, OV798, for colorectal cancer therapy. Mol Cancer Ther 2003; 2: 1003–1009.
Qiu Y, Peng GL, Liu QC, Li FL, Zou XS, He JX . Selective killing of lung cancer cells using carcinoembryonic antigen promoter and double suicide genes, thymidine kinase and cytosine deaminase (pCEA-TK/CD). Cancer Lett 2012; 316: 31–38.
Harris AL . Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer 2002; 2: 38–47.
Vaupel P, Mayer A . Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 2007; 26: 225–239.
Bertout JA, Patel SA, Simon MC . The impact of O2 availability on human cancer. Nat Rev Cancer 2008; 8: 967–975.
Keith B, Johnson RS, Simon MC . HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 2012; 12: 9–22.
Wenger RH, Stiehl DP, Camenisch G . Integration of oxygen signaling at the consensus HRE. Sci STKE 2005; 2005: re12.
Griffiths EA, Pritchard SA, Welch IM, Price PM, West CM . Is the hypoxia-inducible factor pathway important in gastric cancer? Eur J Cancer 2005; 41: 2792–2805.
Rohwer N, Lobitz S, Daskalow K, Jöns T, Vieth M, Schlag PM et al. HIF-1alpha determines the metastatic potential of gastric cancer cells. Br J Cancer 2009; 100: 772–781.
Jung JH, Im S, Jung ES, Kang CS . Clinicopathological implications of the expression of hypoxia-related proteins in gastric cancer. Int J Med Sci 2013; 10: 1217–1223.
Pouysségur J, Dayan F, Mazure NM . Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 2006; 441: 437–443.
Shibata T, Giaccia AJ, Brown JM . Development of a hypoxia- responsive vector for tumor-specific gene therapy. Gene Therapy 2000; 7: 493–498.
Harvey TJ, Hennig IM, Shnyder SD, Cooper PA, Ingram N, Hall GD et al. Adenovirus-mediated hypoxia-targeted gene therapy using HSV thymidine kinase and bacterial nitroreductase prodrug-activating genes in vitro and in vivo. Cancer Gene Ther 2011; 18: 773–784.
Nyati MK, Sreekumar A, Li S, Zhang M, Rynkiewicz SD, Chinnaiyan AM et al. High and selective expression of yeast cytosine deaminase under a carcinoembryonic antigen promoter -enhancer. Cancer Res 2002; 62: 2337–2342.
Liu T, Zhang G, Chen YH, Chen Y, Liu X, Peng J et al. Tissue specific expression of suicide genes delivered by nanoparticles inhibits gastric carcinoma growth. Cancer Biol Ther 2006; 5: 1683–1690.
Koch PE, Guo ZS, Kagawa S, Gu J, Roth JA, Fang B . Augmenting transgene expression from carcinoembryonic antigen (CEA) promoter via a GAL4 gene regulatory system. Mol Ther 2001; 3: 278–283.
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP . Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 2000; 25: 315–319.
Richter AM, Pfeifer GP, Dammann RH . The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta 2009; 1796: 114–128.
Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA . The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol 2002; 22: 4309–4318.
Agathanggelou A, Bièche I, Ahmed-Choudhury J, Nicke B, Dammann R, Baksh S et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res 2003; 63: 5344–5351.
Baksh S, Tommasi S, Fenton S, Yu VC, Martins LM, Pfeifer GP et al. The tumor suppressor RASSF1A and MAP-1 link death receptor signaling to Bax conformational change and cell death. Mol Cell 2005; 18: 637–650.
Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB et al. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem 2006; 281: 4557–4563.
Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res 2006; 66: 2562–2569.
Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell 2007; 27: 962–975.
Dallol A, Agathanggelou A, Tommasi S, Pfeifer GP, Maher ER, Latif F . Involvement of the RASSF1A tumor suppressor gene in controlling cell migration. Cancer Res 2005; 65: 7653–7659.
Wang W, Sun X, Lu L, Zheng JB, Tian Y, Wang W . Cytotoxicity of lymphocytes activated by superantigen toxic-shock-syndrome toxin-1 against colorectal cancer LoVo cells. Mol Cell Biochem 2013; 376: 1–9.
Guo J, Lou W, Ji Y, Zhang S . Effect of CCR7, CXCR4 and VEGF-C on the lymph node metastasis of human pancreatic ductal adenocarcinoma. Oncol Lett 2013; 5: 1572–1578.
Acknowledgements
We thank Center of Translational Medicine, The First Affiliated Hospital, for providing experimental assistance. This work was supported in part by grants from The Natural Science Foundation of China (81172362, 81101874 and 81172359) and the Innovation of Overall Science and Technology Project of Shaanxi province (2013KTCQ03-08). We thank Medjaden Bioscience Limited (Hong Kong, China) for assisting in the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Zhou, PH., Zheng, JB., Wei, GB. et al. Lentivirus-mediated RASSF1A expression suppresses aggressive phenotypes of gastric cancer cells in vitro and in vivo. Gene Ther 22, 793–801 (2015). https://doi.org/10.1038/gt.2015.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2015.49
This article is cited by
-
Suppression of exosomal hsa_circ_0001005 eliminates the Vemurafenib resistance of melanoma
Journal of Cancer Research and Clinical Oncology (2023)
-
The tumor suppressive role of RASSF1A in osteosarcoma through the Wnt signaling pathway
Tumor Biology (2016)