Abstract
Purpose
We evaluated the diagnostic yield in pediatric dilated cardiomyopathy (DCM) of combining exome sequencing (ES)-based targeted analysis and genome-wide copy-number variation (CNV) analysis. Based on our findings, we retrospectively designed an effective approach for genetic testing in pediatric DCM.
Methods
We identified 95 patients (in 85 families) with pediatric onset of DCM. We initially excluded 13 of these families because they already had a genetic diagnosis, leaving a total of 31 probands for single-nucleotide polymorphism (SNP) array and trio-ES. We used Human Phenotype Ontology (HPO)-based filtering for our data analysis.
Results
We reached a genetic diagnosis in 15/31 (48.4%) families. ES yielded a diagnosis in 13 probands (13/15; 86.7%), with most variants being found in genes encoding structural cardiomyocyte components. Two large deletions were identified using SNP array. If we had included the 13 excluded families, our estimated yield would have been 54%.
Conclusion
We propose a standardized, stepwise analysis of (i) well-known cardiomyopathy genes, (ii) CNVs, (iii) all genes assigned to HPO cardiomyopathy, and (iv) if appropriate, genes assigned to other HPO terms. This diagnostic approach yields the highest increase at each subsequent step and reduces analytic effort, cost, the number of variants of unknown clinical significance, and the chance of incidental findings.
Similar content being viewed by others
Introduction
Dilated cardiomyopathy (DCM), characterized by dilation and impaired contraction of the left ventricle or both ventricles, is the most common type of cardiomyopathy (CM) among children younger than 18 years, with an incidence of 0.57 (95% CI: 0.52–0.63) per 100,000 children in the United States.1 The etiology of pediatric DCM encompasses most of the genetic causes that also lead to isolated CM in adults,2,3 and is usually autosomal dominantly (AD) inherited. However, in 14% of pediatric cases, DCM is associated with extracardiac features, pointing toward a wider spectrum of causes, including malformation syndromes, neuromuscular diseases, and metabolic disorders.1 These are often de novo cases or show an X-linked, autosomal recessive (AR) or mitochondrial inheritance pattern. Furthermore, copy-number variants (CNVs) may be identified in patients with multiple congenital anomalies and/or intellectual disability presenting with DCM4,5 as well as in patients with isolated DCM.6,7,8,9
The cause of DCM in children is an independent predictor of the combined outcome of death or transplantation.1 An early genetic diagnosis is therefore very important in determining the etiology, disease course, and prognosis, and may guide rational and personalized treatment choices. In addition, the diagnosis has major implications for family screening and provides insight into the risk of recurrence.
The high degree of genetic heterogeneity in pediatric DCM can currently be addressed via next-generation sequencing (NGS). Most diagnostic labs have developed a targeted NGS panel for adult-onset DCM, but this does not generally encompass the most common and up-to-date neuromuscular, syndromic, and metabolic causes of DCM in childhood. Moreover, the continuous discovery of new disease genes makes it difficult for diagnostic labs to keep up to date on all the genes identified as being involved in pediatric CM; this is a major disadvantage of targeted sequencing using an enriched gene panel. By contrast, exome sequencing (ES), followed by targeted analysis of a regularly updated gene panel, has increasingly become the first-choice approach for heterogeneous diseases (see refs.10,11,12,13 for examples). Pugh et al. showed that the detection rate of pathogenic variants increased from 7–10% to 27–37% by including larger numbers of genes, but there is an increased likelihood of identifying variants of uncertain clinical significance (VUS)2 and of incidental findings (IFs) that do not explain the patient’s phenotype. Human Phenotype Ontology (HPO) has become a useful tool in precision medicine by providing standardized terms to describe phenotypic abnormalities. Each term in the HPO is linked to associated diseases listed in Online Mendelian Inheritance in Man (OMIM) and to related genes by a combination of automated matching of the OMIM Clinical Synopsis to HPO term labels and manual curation of each term.14,15,16,17 These HPO terms are updated monthly and can be used for “personalized” targeted data analysis, thereby reducing the chance of VUS and IFs.
There have been few reports on the diagnostic yield of genetic testing in pediatric DCM. Only one study reported a genetic cause of DCM, in 15 of 41 children (37%); familial disease in this cohort was high (35/41 cases, 85%) and only a limited set of 15 genes was sequenced in 13 patients.18 In Pugh and colleagues’ cohort,2 37% were pediatric DCM cases but an overall diagnostic yield for their pediatric population was not reported. Both studies and current recommendations19,20 endorse genetic testing in the pediatric population. Schedules that define the order of analyses can help reduce the analytical effort needed to detect a genetic cause while minimizing the chance of IFs and the number of VUS. As far as we know, there are no studies reporting the yield and most effective diagnostic approach for genetic testing in patients with childhood-onset DCM.
We therefore had two aims. The first was to evaluate the diagnostic yield of trio-ES-based targeted analysis of genes involved in pediatric DCM in a cohort of thoroughly phenotyped patients, irrespective of their possible disease etiology. Since our current variant-calling pipeline does not detect CNVs (apart from small insertion-deletions), all probands were also subjected to CNV analysis. The second aim was to determine, on the basis of these findings, the optimal diagnostic approach for establishing a genetic diagnosis in as many patients as possible. This involved obtaining the highest increase in yield for each subsequent diagnostic step while minimizing the chance of IFs and the number of VUS, in line with international recommendations.20
Materials and methods
Patient selection
The University Medical Centre Groningen’s (UMCG’s) medical research ethics committee approved this study (approval no. 2014092), and written informed consent was obtained from all participants or their legal representatives.
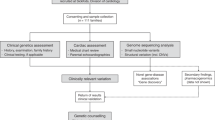
We identified 95 patients (from 85 families) with familial and nonfamilial DCM or a mixed cardiac phenotype with age at onset of CM <18 years. These included patients with extracardiac features or possible myocarditis, or childhood cancer survivors who developed DCM after anthracycline treatment, as well as patients who experienced partial recovery from their DCM (Supplementary Tables S1–S3 online). All patients were referred to either UMCG or Leiden University Medical Centre (LUMC), The Netherlands, between May 1993 and April 2017 for genetic counseling. DCM was defined by the presence of left ventricle (LV) dilatation (LV end-diastolic dimension >2 SD above mean for body surface area) and systolic dysfunction (fractional shortening or LV ejection fraction >2 SD below mean for age) not explained by abnormal loading conditions20 or evidence for DCM from autopsy.
Patients with a genetic diagnosis explaining their phenotype and the course of their disease were excluded from our ES and CNV analysis for this project (Supplementary Table S1). We did, however, offer ES to patients with a (likely) pathogenic variant that may not fully explain their phenotype or disease course, for example, a 9-year-old patient with a dramatic disease course who carried a truncating TTN variant, which is usually associated with adult onset and a relatively benign course.21 We recontacted all the remaining gene/mutation-elusive DCM patients who had been evaluated before the introduction of diagnostic ES.
All patients were phenotyped by a cardiologist and clinical geneticist, sometimes accompanied by a pediatric neurologist, all of whom reviewed the case. The clinical geneticist examined the patient to look for dysmorphisms and extracardiac features at the time of counseling for ES and CNV analysis and took a three-generation family history. and physically examined the patient to look for dysmorphisms and neurological and other extracardiac features at the time of counseling for ES and CNV analysis.
CNV analysis and homozygosity mapping (single-nucleotide polymorphism (SNP) array)
Genomic DNA from the affected child and its parents was extracted from peripheral blood or fibroblasts. Genome-wide genotyping, using HumanCytoSNP-850K SNP array according to the manufacturer’s protocols (Illumina, San Diego, CA), was performed to identify CNVs and/or copy-number neutral homozygous regions, as described in Supplementary Materials and Methods.
Exome sequencing and data analysis
Exome sequencing and variant calling are described in the Supplementary Materials and Methods. Briefly, the exome was captured with the Agilent Sureselect XT Human All Exon V5 or V6 kit (Agilent, Santa Clara, CA). Exome libraries were sequenced on an HiSeq2500 or a NextSeq500 machine (Illumina, San Diego, CA) with 2 × 100 bp and 2 × 150 bp paired-end reads, respectively, at an average coverage of 100 × and with >90% of the exome covered >20 ×. Sequence reads were aligned to the human reference genome GRCh37/hg19 with the Burrows-Wheeler Aligner version 0.7.5a.22 Variants were called using Genome Analysis Toolkit (GATK) software.23,24 For two patients (F6P1 and F26P1), whole-genome sequencing (WGS) was performed to avoid time-consuming capturing steps because of their severe clinical conditions and young age, as described previously25 (Supplementary Materials and Methods).
A gene list of 310 genes was created based on the HPO term cardiomyopathy (HP:0001638; http://www.human-phenotype-ontology.org/), supplemented with genes not included in this HPO term but associated with CM in recent scientific reports26 (Supplementary Table S4). To generate a “private” gene panel, this gene list was expanded with genes assigned to other HPO terms for individual patients when appropriate (Supplementary Table S5). In order to define the most relevant HPO terms, a clinical geneticist discussed the patients’ phenotypes with a pediatric cardiologist, a pediatrician, a neurologist, and/or other relevant physicians.
Potentially relevant variants were evaluated in a multidisciplinary meeting with at least a clinical geneticist, a molecular geneticist, and a laboratory technician present. When necessary, a cardiologist, a neurologist, and/or a pediatrician were also consulted. An independent expert panel was set up to discuss the clinical relevance of IFs, predefined as likely or definitely pathogenic variants in known disease genes unrelated to the patient’s current phenotype, as described elsewhere.25
Calculation of internal and potential diagnostic yield
The internal yield was calculated as the number of families in which a genetic diagnosis was found divided by the total number of families subjected to CNV analysis and ES. Since we excluded 13 families whose genetic diagnosis had been established by other techniques (multiplex ligation-dependent probe amplification (MLPA), Sanger sequencing, or targeted NGS), the internal yield is an underestimation of the yield in an unselected cohort. To adjust for this, these 13 families were included in our analysis retrospectively.
The potential yield, assuming that all 85 families were offered ES/CNV analysis as routine genetic testing, was estimated as the internal yield plus a proportion of the 13 families who were not included initially (because in this group too, a subset of patients may not respond to the invitation or may refrain from further testing). We applied a correction factor of 0.43 (proportion of families subjected to ES/SNP array divided by the total number of families eligible for ES/CNV analysis; 31/72). The 95% confidence intervals of the potential yields were calculated as exact binomial confidence intervals.
Results
The clinical and genetic results for all 35 patients are summarized in Tables 1Tables 2Tables 3 and Supplementary Tables S1,S2, and S6.
Subjects and clinical characteristics
Figure 1 depicts the study setup. After excluding 16 patients (13 families) who already had a genetic diagnosis that explained their phenotype and young age at onset (Supplementary Table S1), we identified 79 patients from 72 families that fulfilled our inclusion criteria. For 19 families, either no DNA or tissue for a deceased patient was available for testing or it was not feasible to inform the patients/families about new diagnostic possibilities (i.e., NGS); another 14 patients (14 families) did not respond to the invitation, and 9 patients (8 families) refrained from further testing. We performed trio-based ES (WGS in two) and CNV analysis in 31 unrelated families (31 probands and 4 siblings in total; patient characteristics are shown in Table 1 and Supplementary Table S2).
Study profile and diagnostic yield. We could not reach or test 19 families because, for example, an address was unavailable or the patient had died and no DNA/tissue was available. Of the 72 families who initially had no genetic diagnosis, 31 (43%) were subjected to ES/CNV analysis. Characteristic features of mitochondrial disease were reviewed by Leonard and Schapira.40 ES/CNV, exome sequencing/copy-number variation; mtDNA, mitochondrial DNA; ES/SNP, exome sequencing/single-nucleotide polymorphism.
Internal yield
A genetic diagnosis could be made in 15 of 31 families (48.4%, 95% CI: 30.2–66.9%) (Table 2). In 14 families (45.2%), the identified genetic defect could explain their DCM. The mutation in the 15th family has not been associated with DCM so far, but it does explain additional clinical features seen in the patient (see also below and Supplementary Data 2). Exome sequencing yielded a diagnosis in 13/15 families (86.7%). Except for one, these pathogenic or likely pathogenic variants were identified in eight CM-associated genes. Ten of the 13 variants (76.9%) were heterozygous and found in genes encoding structural components of cardiomyocytes, two of these were de novo (Table 2).
In 2/13 (15.4%) families diagnosed by ES, we identified causal variants in less-well-known CM genes: one homozygous variant in the beta-1 galactosidase (GLB1) gene and one homozygous variant in the SPEG complex locus (SPEG) gene. In one patient (F6P1) carrying the GLB1 mutation, we found an additional likely pathogenic variant in the T-box 20 (TBX20) gene. The probands in both these families had consanguineous parents and showed extracardiac features (Table 2).
Expanding our virtual gene panel by adding HPO terms to the filtering tree in patients with additional features identified compound heterozygous variants c.874C>T, p.(Arg292*) and c.3118_3121delAACA, p.(Asn1040Glufs*9) in CEP135 in patient F25P1. These explain her microcephaly and developmental delay, but it is unclear whether they also explain her DCM (for further discussion, see Supplementary Data).
In addition to the 13 families diagnosed by ES, two diagnoses were made by SNP array (2/15; 13.3%, Table 2). In patient F29P1, who presented with a severely dilated left ventricle, several apical VSDs, and an open ductus arteriosus at age 2 weeks, a de novo 1p36.33p36.32 deletion was identified consistent with chromosome 1p36 deletion syndrome (MIM607872). In patient F31P1, who presented with decompensated heart failure at age 16 years, a de novo 10q25.2 deletion was identified encompassing the RNA-binding motif protein 20 (RBM20) gene. A deletion including this gene has not been described before, although RBM20 missense variants have been associated with early-onset DCM, end-stage heart failure, and high mortality;27 several studies suggest a loss-of-function effect.28,29
Importantly, we determined a genetic diagnosis for 6/13 patients (46.1%) with possible acute myocarditis. Although they did not meet the Dallas criteria,30 they suffered from prodromal symptoms mimicking a viral infection, including abdominal pain, vomiting, upper respiratory tract infections, and coughing. We further reached a genetic diagnosis in 1/2 patients treated with anthracycline.
Potential yield
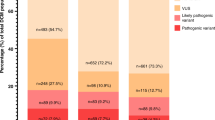
Because we excluded patients with a known genetic diagnosis, our overall diagnostic yield is an underestimation. To compensate, we included these families retrospectively and calculated the potential yield. If we had included the 13 families who were initially excluded, the estimated yield would have been 53.9% (95% CI: 37.4–68.7%) (Figure 2).
Our proposed stepwise diagnostic approach for analyzing pediatric DCM patients and the resulting internal and potential yields. Step 1. Analysis of a limited selection of 60 genes, encoding mostly structural components of cardiomyocytes. Step 2. CNV analysis, including homozygosity analysis. Step 3. Reanalysis of the data by filtering for variants in a comprehensive virtual gene panel, based on the HPO term “cardiomyopathy.” Step 4. Adding more HPO terms for filtering in those patients presenting with additional cardiac and/or noncardiac features. In the group without a diagnosis, 43% (31/72 families) agreed to undergo ES and CNV analysis. We reasoned that the group of 16 patients (from 13 families, Supplementary Table S1) with a known genetic diagnosis, to whom we did not offer ES/CNV analysis, would not differ much from the group that was eligible for ES/CNV analysis (e.g., no bias in severity of DCM or number of familial cases). Therefore, the same percentage of inclusions should also be applicable to this “not offered ES/CNV analysis” group. Eleven of these 13 families would have been identified by our approach if ES/CNV analysis had been performed as the first-tier test. The potential yield would therefore be the sum of the internal yield (15/31 families) plus the 0.43 proportion of the excluded patients who already had a genetic diagnosis (15+(11*0.43))/(31+(13*0.43)), which is 53.9%. CM, cardiomyopathy; CNV, copy-number variation; DCM, dilated cardiomyopathy; ES, exome sequencing; HPO, Human Phenotype Ontology; SNP, single-nucleotide polymorphism; VUS, variant of uncertain clinical significance; WES, whole-exome sequencing.
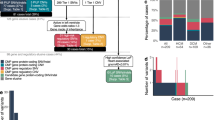
Variants of unknown significance
We identified up to 11 VUS per patient (n = 96, mean 2.74 per patient), including 5 chromosomal CNVs (Tables 2 and 3). During our study, the MYL2 c.263A>C, p.(Glu88Ala) variant (identified in two patients) was reclassified as likely pathogenic, based on segregation analysis in three families and the identification of a common haplotype (Supplementary Data 1 and Supplementary Figure S1).
Incidental findings
No IFs in AD inherited disease genes were found. A pathogenic variant in AR inherited disease genes was identified in 5 of the 35 patients (14.3%) (Tables 2 and 3): in GYS1 (Glycogen storage disease 0, muscle, OMIM 138570), in SGSH (Mucopolysaccharidosis type IIIA, OMIM 252900), in PEX1 (peroxisome biogenesis disorder 1A, OMIM 214100), in PMM2 (congenital disorder of glycosylation type Ia, OMIM 212065), and in RAB3GAP2 (Warburg micro syndrome 2, OMIM 614225; Martsolf syndrome, OMIM 212720). None of the patients’ phenotypes fits the respective disease, and the carrier frequency of a disease-causing mutation in each of the five genes is <1/60.
Toward the most effective analysis strategy
Based on our results, we retrospectively tested a stepwise analysis on our data. To determine the most effective approach for genetic testing and data analysis, leading to the highest increase in yield at each subsequent diagnostic step, we also included the patients who already had a diagnosis. Most such genetic diagnoses would have been made by analyzing only a limited selection of well-known CM genes (35.6%; 95% CI: 22.7–53.0%) (Figure 2, step 1). Subsequently, adding CNV analysis would have resulted in an increase in diagnostic yield to 44.6% (95% CI: 29.9–61.1%) (Figure 2, step 2). Finally, filtering the data by generating a virtual gene panel based on the HPO-term cardiomyopathy (HP:0001638) in the patients still without a genetic diagnosis and subsequently adding more HPO terms, where appropriate, would have led to an increase of up to 51.2% (95% CI: 34.8–66.2%) and 53.9% (95% CI: 37.4–68.7%) diagnoses, respectively.
The number of VUS identified per step are shown in Figure 2. We only encountered carriers of AR diseases at step 3 in this study and no IFs.
Discussion
Pediatric DCM is a relatively rare but life-threatening disease with a strong genetic component. Genetic testing improves its clinical management and has become an essential part of contemporary care for children with DCM and their families. NGS technology offers excellent opportunities for efficient genetic diagnostics in general, but little has been reported for pediatric DCM. We had a relatively large cohort of unselected, well-phenotyped patients that provided a unique opportunity to implement and test the efficacy of an approach involving CNV analysis and trio-based ES with analysis of a virtual gene panel of more than 310 CM-related genes. The use of HPO terms for data filtering has major advantages over a fixed gene panel as they are automatically updated and patient-specific terms can be used. Testing 31 probands with DCM and their parents, our approach yielded a diagnosis in 48.4% (15/31). Applying this approach retrospectively to all our cases, including those who already had a genetic diagnosis, would have resulted in a yield >50%.
We have also shown that the same genetic workup is justified in patients with possible myocarditis and childhood cancer survivors in whom DCM presents as a toxic side effect. The high yield obtained with our approach also holds an important implication: that in many cases, a “genetics-first” approach can reduce or even avoid costly intensive care for some children (e.g., F6P1 and F29P1) or replace other time-consuming and/or invasive, nongenetic diagnostic testing (e.g., F7P1). Moreover, a prospective study by Stark et al.31 showed that ES used early in the diagnostic pathway more than triples the diagnostic rate for one-third of the cost per diagnosis.
Stepwise diagnostic approach
To reduce the analytic effort, the number of VUS, the cost, and the chance of identifying IFs, we now propose a standardized, stepwise analysis of a subset of genomic data in pediatric DCM cases (Figure 2). We prioritized the diagnostic steps according to the highest increase in yield per step. We do not claim that this strategy provides the best or quickest analytical path in general; rather, it represents an ad hoc approach inferred from our current results. The generalizability of our stepwise approach needs to be corroborated in larger cohorts, which also may lead to obtain greater insight into the meaning of variants in genes that are less often associated with CM.
Our study shows that restricting the analysis to genes encoding desmosomal cell adhesion proteins, sarcomeric proteins, and a few other, well-established CM genes (n = 60) would potentially yield a diagnosis in 35.6% of pediatric patients. This is slightly lower than the 37% yield in a pediatric cohort reported by Rampersaud et al.18 However, they included predominantly familial DCM (85% of cases), whereas we included both familial cases (23%) and patients with a negative family history for CM (77%), although cardiac screening was performed in most, but not all, first-degree relatives.
The addition of a CNV analysis would increase our potential yield to 44.6%. This rise can be explained by the relatively high percentage of patients diagnosed with Duchenne muscular dystrophy (Supplementary Table S1) who carry a deletion/duplication of (at least) the DMD gene.
Finally, reanalyzing the data by filtering for variants in a comprehensive, but personalized, virtual gene panel based on HPO cardiomyopathy, plus additional HPO terms where appropriate, would have resulted in a total diagnostic potential of 51.2% and 53.9%, respectively (Figure 2, steps 3 and 4). Therefore, the presence/absence of syndromic or extracardiac features should indicate the extent of genetic analysis required in a clinical setting. The use of HPO terms and stepwise data analysis reduces the number of VUS and the chance of identifying IFs, which is in accordance with current guidelines for genetic testing in minors32 and more specifically for DCM.19
Pros and cons of our ES-based approach
Currently, the strategies for NGS of patients with CM can be divided into two fundamentally different approaches: targeted gene panel sequencing, with capture of a restricted set of genes, and ES/WGS. The pros and cons of these approaches for other patient categories have been extensively reviewed.33 Our approach combines the following advantages of both methods (for related examples from our cohort, see Supplementary Data 2).
-
1.
Although ES data interpretation is, in general, more complicated and time-consuming than targeted gene panel sequencing, using virtual gene panels to filter ES data reduces the number of genetic variants needing to be interpreted. Moreover, by filtering for variants in the genes of interest, the number of IFs and VUS is reduced.
-
2.
In consanguineous families, variant filtering may first be restricted to homozygous regions.
-
3.
Multiple relevant variants in one patient can be detected.18,34,35,36
-
4.
Novel traits may be identified by using additional HPO terms.
-
5.
Our filter strategy can be applied to all cardiomyopathies that show clinical and genetic overlap and that are sometimes difficult to classify on echocardiographic parameters.
-
6.
Having the full exome available offers the option to broaden the analysis if a diagnosis cannot be made and/or if clinical development indicates that other approaches are needed. This may involve alternative filtering or “opening” the exome, which could lead to identification of novel genes associated with pediatric CM, as we recently showed for ALPK3.37
However, an ES-based diagnostic approach also has some disadvantages:
-
1.
A potentially lower vertical coverage of some regions compared to targeted gene panel sequencing may lead to some mutations being missed, although LaDuca et al.38 showed that ES covered 99.6% of mutations identified on a targeted NGS panel for cardiovascular diseases. Nevertheless, in patients for whom a specific disease, such as DMD, is strongly suspected, targeted sequencing of the candidate gene, including MLPA, may be preferred to ES.
-
2.
Broadening analysis to a larger gene panel runs the risk of identifying VUS, as seen by the 2.5-fold higher yield of VUS when we used expanded gene panels in our study. However, ES does allow for stepwise data analysis to minimize the number of additional VUS. The accuracy of variant interpretation can be improved by access to larger libraries of comprehensive sequencing data in well-phenotyped CM patients and well-documented controls, and with the availability of affected family members for segregation analyses.
-
3.
A longer turnaround time. During our study, turnaround time was cut from >26 weeks to 6 weeks, with the potential for even shorter times.
-
4.
ES-based analysis is more expensive than targeted NGS. However, continuous technological innovations have already reduced its cost, and this trend will continue.
-
5.
Exome sequencing cannot be used to detect mutations in noncoding regions or in mitochondrial DNA (mtDNA), and, at the moment, our ES pipeline is not able to detect CNVs. However, several algorithms have been developed using a read-depth approach to identify CNVs in ES data,39 which could increase our yield further.
WGS has several advantages over ES, including the possibility of detecting mtDNA sequence variants and CNVs and the elimination of time-consuming capturing steps, which is important for critically ill patients for whom a rapid diagnosis may influence treatment options.25 WGS may therefore be preferred over combined ES/CNV analysis in the future. However, given the extra cost of WGS, ES followed by virtual targeted stepwise analyses is currently still our preferred approach in a clinical diagnostic setting.
Future work and perspectives
We are now analyzing the full exomes for our cohort in a research setting, using a gene-prioritization method based on gene coexpression networks. We are also exploring the possibility of parallel ES and RNA sequencing to help pinpoint candidate genes and mutations. Careful family and cascade screening will offer more insight into the putative pathogenesis of VUS, while data sharing, mining, and curation, in vitro and in vivo functional analyses, RNA-sequencing studies, and metabolomics are essential to gaining more insight into the pathogenicity of novel variants in genes implicated in pediatric DCM.
Conclusion
We have demonstrated that combining CNV analysis with trio-based ES yields a fast diagnosis in more than 50% of pediatric DCM probands, including those with possible myocarditis. To reach the highest increase in yield and reduce the chance of detecting noncontributory VUS or IFs, we propose a standardized, stepwise analysis of (i) well-known CM genes, (ii) CNVs, (iii) all genes assigned to HPO cardiomyopathy, and (iv) if appropriate, genes assigned to other HPO terms in pediatric DCM cases. Stepwise filtering for variants in a virtual and flexible gene panel based on HPO terms is an effective diagnostic strategy for pediatric DCM. We report on a relatively small patient cohort, owing to the rarity of this disorder. However, the relevance of our approach should be explored in other and/or larger patient cohorts to confirm the superiority of CNV analysis and ES combined with targeted data analysis over other molecular and bioinformatic diagnostic approaches. This also applies to the proposed strategy for adult DCM/CM patients, in whom an AD mode of inheritance is more prevalent than in children and in whom the phenotype has become clearer regarding noncardiac symptoms. The need for rapid diagnosis is generally less urgent in adults than in children. DCM in children may demand a different genetic approach from that used for adults.
References
Towbin JA, Lowe AM, Colan SD et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 2006;296:1867–1876.
Pugh TJ, Kelly MA, Gowrisankar S et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med 2014;16:601–608.
Posafalvi A, Herkert JC, Sinke RJ et al. Clinical utility gene card for: dilated cardiomyopathy (CMD). Eur J Hum Genet 2013;21.
Battaglia A, Hoyme HE, Dallapiccola B et al. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 2008;121:404–410.
Shoukier M, Schroder J, Zoll B et al. A de novo interstitial deletion of 2p23.3-24.3 in a boy presenting with intellectual disability, overgrowth, dysmorphic features, skeletal myopathy, dilated cardiomyopathy. Am J Med Genet A 2012;158A:429–433.
Norton N, Li D, Rieder MJ et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet 2011;88:273–282.
Cohen N, Muntoni F. Multiple pathogenetic mechanisms in X linked dilated cardiomyopathy. Heart 2004;90:835–841.
Gupta P, Bilinska ZT, Sylvius N et al. Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol 2010;105:365–377.
Ceyhan-Birsoy O, Pugh TJ, Bowser MJ et al. Next generation sequencing-based copy number analysis reveals low prevalence of deletions and duplications in 46 genes associated with genetic cardiomyopathies. Mol Genet Genomic Med 2015;4:143–151.
Wortmann SB, Koolen DA, Smeitink JA, van den Heuvel L, Rodenburg RJ. Whole exome sequencing of suspected mitochondrial patients in clinical practice. J Inherit Metab Dis 2015;38:437–443.
Willemsen MH, Kleefstra T. Making headway with genetic diagnostics of intellectual disabilities. Clin Genet 2014;85:101–110.
Visser G, Suormala T, Smit GP et al. 3-methylcrotonyl-CoA carboxylase deficiency in an infant with cardiomyopathy, in her brother with developmental delay and in their asymptomatic father. Eur J Pediatr 2000;159:901–904.
Neubauer J, Lecca MR, Russo G et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet 2017;25:404–409.
Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008;83:610–615.
Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet 2010;77:525–534.
Fang H, Wu Y, Yang H et al. Whole genome sequencing of one complex pedigree illustrates challenges with genomic medicine. BMC Med Genomics 2017;10.
Kohler S, Vasilevsky NA, Engelstad M et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res 2017;45:D865–D876.
Rampersaud E, Siegfried JD, Norton N, Li D, Martin E, Hershberger RE. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog Pediatr Cardiol 2011;31:39–47.
Pinto YM, Elliott PM, Arbustini E et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J 2016;37:1850–1858.
Elliott P, Andersson B, Arbustini E et al. Classification of the cardiomyopathies: a position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2008;29:270–276.
Jansweijer JA, Nieuwhof K, Russo F et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur J Heart Fail 2017;19:512–521.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.
McKenna A, Hanna M, Banks E et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Van der Auwera GA, Carneiro MO, Hartl C et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43.
van Diemen CC, Kerstjens-Frederikse WS, Bergman KA et al. Rapid targeted genomics in critically ill newborns. Pediatrics 2017;140.
Harakalova M, Kummeling G, Sammani A et al. A systematic analysis of genetic dilated cardiomyopathy reveals numerous ubiquitously expressed and muscle-specific genes. Eur J Heart Fail 2015;17:484–493.
Brauch KM, Karst ML, Herron KJ et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol 2009;54:930–941.
Guo W, Schafer S, Greaser ML et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med 2012;18:766–773.
Beqqali A, Bollen IA, Rasmussen TB et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc Res 2016;112:452–463.
Aretz HT, Billingham ME, Edwards WD et al. Myocarditis. A histopathologic definition and classification. Am J Cardiovasc Pathol 1987;1:3–14.
Stark Z, Schofield D, Alam K et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med 2017;19:867–874.
Botkin JR, Belmont JW, Berg JS et al. Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am J Hum Genet 2015;97:6–21.
Carroll CJ, Brilhante V, Suomalainen A. Next-generation sequencing for mitochondrial disorders. Br J Pharmacol 2014;171:1837–1853.
Millat G, Bouvagnet P, Chevalier P et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur J Med Genet 2011;54:e570–5.
Roncarati R, Viviani Anselmi C, Krawitz P et al. Doubly heterozygous LMNA and TTN mutations revealed by exome sequencing in a severe form of dilated cardiomyopathy. Eur J Hum Genet 2013;21:1105–1111.
Golbus JR, Puckelwartz MJ, Dellefave-Castillo L et al. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ Cardiovasc Genet 2014;7:751–759.
Almomani R, Verhagen JM, Herkert JC et al. Biallelic truncating mutations in ALPK3 cause severe pediatric cardiomyopathy. J Am Coll Cardiol 2016;67:515–525.
LaDuca H, Farwell KD, Vuong H et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One 2017;12:e0170843.
Pfundt R, Del Rosario M, Vissers LELM et al. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med 2017;19:667–675.
Leonard JV, Schapira AH. Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet 2000;355:299–304.
Acknowledgments
We are grateful to the patients and families for their participation. We thank Jackie Senior for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Herkert, J.C., Abbott, K.M., Birnie, E. et al. Toward an effective exome-based genetic testing strategy in pediatric dilated cardiomyopathy. Genet Med 20, 1374–1386 (2018). https://doi.org/10.1038/gim.2018.9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2018.9
Keywords
This article is cited by
-
The role of TBX18 in congenital heart defects in humans not confirmed
European Journal of Human Genetics (2023)
-
Genetic variant burden and adverse outcomes in pediatric cardiomyopathy
Pediatric Research (2021)
-
Recessive ciliopathy mutations in primary endocardial fibroelastosis: a rare neonatal cardiomyopathy in a case of Alstrom syndrome
Journal of Molecular Medicine (2021)
-
De novo variants in CAMTA1 cause a syndrome variably associated with spasticity, ataxia, and intellectual disability
European Journal of Human Genetics (2020)
-
Advantages and Perils of Clinical Whole-Exome and Whole-Genome Sequencing in Cardiomyopathy
Cardiovascular Drugs and Therapy (2020)