Abstract
Purpose
Great uncertainty exists about the costs associated with whole-genome sequencing (WGS).
Methods
One hundred cardiology patients with cardiomyopathy diagnoses and 100 ostensibly healthy primary care patients were randomized to receive a family-history report alone or with a WGS report. Cardiology patients also reviewed prior genetic test results. WGS costs were estimated by tracking resource use and staff time. Downstream costs were estimated by identifying services in administrative data, medical records, and patient surveys for 6 months.
Results
The incremental cost per patient of WGS testing was $5,098 in cardiology settings and $5,073 in primary care settings compared with family history alone. Mean 6-month downstream costs did not differ statistically between the control and WGS arms in either setting (cardiology: difference = −$1,560, 95% confidence interval −$7,558 to $3,866, p = 0.36; primary care: difference = $681, 95% confidence interval −$884 to $2,171, p = 0.70). Scenario analyses showed the cost reduction of omitting or limiting the types of secondary findings was less than $69 and $182 per patient in cardiology and primary care, respectively.
Conclusion
Short-term costs of WGS were driven by the costs of sequencing and interpretation rather than downstream health care. Disclosing additional types of secondary findings has a limited cost impact following disclosure.
Similar content being viewed by others
Introduction
Whole-genome sequencing (WGS) can facilitate molecular diagnoses and identify genetic variants to characterize disease risks, tailor medications, screen for recessive traits, and more. Dramatic improvements in its cost, speed, and capabilities are fueling expectations that WGS will become an important part of everyday patient care,1,2 and some commentators hope that it will streamline diagnoses and enhance disease prevention.3 Early evidence is promising,4,5 but there are concerns that WGS may also initiate a cascade of confirmatory testing and ongoing screening that greatly increases health-care expenditures.6,7 Empirical data to inform the discussion are sparse.
Studies to date have examined the costs of genomic sequencing to provide molecular diagnoses for specific syndromes8,9,10,11,12,13 or have projected the potential cost impact of disclosing a limited set of secondary findings.14 However, these early attempts do not fully account for more extensive secondary findings such as pharmacogenomic applications and risk predictions for common disease. Furthermore, current modeling efforts assume rational responses despite evidence that patients often respond to secondary findings in unexpected ways15,16 and despite evidence that physicians are often unclear about their obligations to act on information that is unrelated to the primary indication for testing.17 Few clinical studies have examined the costs of integrating WGS into the everyday care of patients, including broader applications that capitalize on the full potential of WGS.
To fill these knowledge gaps, we conducted a cost analysis of the MedSeq Project, a pair of randomized controlled trials of WGS in cardiology and primary care settings. Limited prior analyses within the primary care setting revealed some evidence for increased health-care utilization and costs within the Partners HealthCare system following WGS.18 The analyses presented here extend these findings through a more comprehensive accounting of in- and out-of-system health-care services and out-of-pocket costs in both cardiology and primary care settings, including a microcosting analysis of WGS itself and scenario and sensitivity analyses that examine the impact of different reporting strategies and cost assumptions. The objective of these analyses is to provide novel insight about the short-term cost impact of integrating WGS into medical care.
Materials and methods
Clinical trial overview
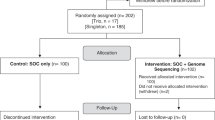
The rationale, design, and primary molecular and clinical findings of the MedSeq Project were previously reported,17,18,19,20,21,22,23 and methodological details are summarized in detail in Supplementary Appendix S1 online. Briefly, primary care physicians identified ostensibly healthy patients between 40 and 65 years of age and cardiologists identified adult patients of any age with diagnoses of hypertrophic or dilated cardiomyopathy (HCM/DCM) for recruitment by MedSeq Project staff. To compare panel-based genetic testing with WGS in patients with HCM/DCM, cardiology patients were either confirmed to have had panel testing or underwent panel testing prior to MedSeq Project disclosure sessions. At their first study visit, patients consented for participation, provided family-history information, had blood drawn, and provided baseline information. Using concealed envelopes, study staff then randomly assigned patients to either a “control arm” that received a structured family-history review or a “WGS arm” that received family history plus a WGS analysis and report. Cardiologists also reviewed their patients’ prior genetic test results in both randomization arms.
WGS reports included findings related to the patients’ condition (cardiology patients only), monogenic disease risks associated with well-established genetic conditions, carrier status for autosomal recessive conditions, pharmacogenomic findings, and risk predictions based on markers for cardiometabolic traits.19,20,22 Prior to disclosure, physicians could contact a Genome Resource Center staffed by genetic counselors and medical geneticists to ask questions about reports or study procedures. Investigators were not blinded to randomization, although patient participants were blinded until disclosure sessions and physician participants were blinded until they received family-history and WGS reports, if applicable. The Partners Human Research Committee and Baylor College of Medicine Institutional Review Board both approved the study protocol (ClinicalTrials.gov NCT01736566). The protocol, statistical code, and data set are available upon request from the corresponding author. Genetic data are available via dbGaP (accession number phs000958.v1.p1).
Cost analysis
Overall approach
We undertook a planned cost analysis alongside the pilot randomized controlled trial using guidelines published by the International Society for Pharmacoeconomics and Outcomes Research and the Second Panel on Cost-Effectiveness in Health and Medicine.24,25 Costs are presented in 2015 US dollars.
Costs of WGS
To understand the costs of WGS to the health-care system, we used a microcosting approach with program operation logs,26 in which staff measured personnel effort and resource use using a combination of forms that tracked each DNA sample and by reviewing audiorecorded consent and disclosure sessions to determine their lengths. Consent sessions for MedSeq Project participation were assumed to be comparable in length to consent sessions for WGS. Personnel effort was translated to costs using 2015 wage data as described in more detail in Supplementary Appendix S4 online. We assigned costs for disclosure sessions based on Centers for Medicare and Medicaid Services (CMS) fee schedules for outpatient evaluation and management visits. The cost of sequencing was estimated from data provided by the National Human Genome Research Institute,27 and costs for confirming monogenic and carrier findings via Sanger sequencing were based on 2015 Laboratory for Molecular Medicine rates for familial variant testing.
Downstream health-care utilization and costs
To understand the short-term impact of WGS disclosure on follow-up health-sector spending, we assessed costs over the 6 months preceding results disclosure and the 6 months following disclosure for both randomization arms. We compared postdisclosure costs by randomization status, as well as separately comparing whether the difference in predisclosure and postdisclosure costs differed by randomization status. We supplemented health-care services identified in administrative data accessed via the Partners HealthCare Research Patient Data Registry with services identified through a review of medical records. We also incorporated services reported in patient surveys completed at disclosure sessions and 6 weeks and 6 months postdisclosure that were not identified in the registry or medical records data. Costs for services were estimated by multiplying utilization by cost weights derived from CMS fee schedules as described in Supplementary Appendix S4 online. Patient out-of-pocket expenses during the postdisclosure period were assessed using survey items adapted from the McMaster Cost of Care Questionnaire,28 where patient participants reported average monthly costs for procedural and visit copays, medical equipment, medications, and additional expenses related to their health care. Differences from prior-reported costs18 are due primarily to the addition of services identified through patient self-report.
Sensitivity and scenario analyses
Sensitivity and scenario analyses incorporating both WGS costs and downstream costs examined how findings varied with different analytic assumptions. First, we examined how costs changed when different types of downstream health-care services were included. One approach examined “immediately attributable” services, including only services that physicians reported recommending in response to family-history review and/or family-history review and WGS reports, and that were confirmed through medical record review or patient self-report. Another approach included estimates of the costs for a comprehensive workup of monogenic disease risks per guidelines such as GeneReviews, clinical synopses such as OMIM, and published medical literature.29,30 We also examined the implications of different reporting criteria, such as omitting carrier status or polygenic risk predictions, and reporting no secondary findings or reporting only secondary monogenic disease risk and carrier findings classified as likely pathogenic or pathogenic, per American College of Medical Genetics and Genomics recommendations.31 In addition, we varied cost assumptions of WGS to reflect the range of prices available at CLIA-certified labs, and we varied health-care price weights to be 50 to 200% of CMS fee schedule amounts. Finally, we analyzed costs from a third-party payer perspective by omitting patient out-of-pocket expenses.
Statistical approach
A priori enrollment goals were not established given that this was a pilot study. We used multivariable linear regression to compare arithmetic mean costs by randomization arms, as recommended,24 including all observed data and imputing data for missing observations. Models included terms for WGS versus control randomization status, cardiology versus primary care, an interaction term, and baseline health status. Covariates were included if they improved model precision (i.e., p < 0.05). We also used this approach to compare the difference in health-care spending in the 6-month period postdisclosure against the 6-month period predisclosure, but omitted patient out-of-pocket expenses because relevant survey data were not available. Subanalyses included additional terms as relevant to examine changes over time, to examine the impact of learning about an unexpected monogenic disease risk, or to examine the impact of cardiometabolic risk predictions. Although terms for interactions between cohorts and randomization status were not significant, we report cohorts separately because downstream costs among cardiology patients were much larger than downstream costs among primary care patients. To compare changes in health-care utilization, we used nonparametric tests (Wilcoxon rank-sum and Wilcoxon signed-rank tests) because data were highly skewed.
Four randomized individuals who did not attend disclosure sessions were omitted from analyses (Supplementary Appendix S5 online). Analyses were conducted using R version 3.4.3. As recommended for smaller samples, confidence intervals were calculated using nonparametric bootstrapping (1,000 samples).24 We assumed that missing survey data were missing at random, and imputed nonresponse with fully conditional specification, running 20 iterations to create each of 20 imputed data sets for each bootstrapped sample. Imputed data accounted for less than 1% of WGS costs through disclosure and 1.2% of downstream medical costs.
Results
Sample characteristics and WGS findings
Two hundred participants attended results disclosure sessions. Demographic characteristics did not vary by randomization status except on educational attainment among primary care patients (Table 1). Table 2 summarizes monogenic and carrier findings among patients who received WGS. All cardiomyopathy variants that had been identified during panel testing of cardiology patients were confirmed by WGS, although an 18–base pair duplication in MYBPC3 may have been missed if investigators did not know to look for it because it had been identified during prior targeted HCM genetic testing. In addition, among the 22 cardiology patients randomized to WGS who had no variants identified in panel testing, a pathogenic variant associated with HCM was identified in 1. Of the 27 cardiology patients who had positive or inconclusive panel testing results, 3 with inconclusive prior results were identified with a previously unreported variant of uncertain significance associated with their DCM or HCM diagnosis. Secondary findings about monogenic disease risks and carrier status for autosomal recessive conditions were identified in 8 (16%) and 41 (84%) cardiology patients in the WGS arm, respectively.
Among the 50 primary care patients who received WGS, we identified variants associated with monogenic disease in 13 (26%), although 2 patients who received information about hereditary hemochromatosis had already been diagnosed with the condition. All primary care patients in the WGS arm (100%) received information about carrier status for autosomal conditions. Specific variants are detailed in Supplementary Appendix S6 (monogenic disease risk findings) and Supplementary Appendix S7 online (carrier findings).
As noted previously, all cardiology and primary care patients in the WGS arms also received polygenic risk predictions for eight cardiometabolic traits and pharmacogenomic information for five drugs, summarized in Supplementary Appendix S8 online.
Cost of sequencing
Table 3 summarizes the time demands and associated costs of family-history review and WGS reporting. The average incremental cost per patient for WGS, including variant interpretation and disclosure, was $5,098 in cardiology settings and $5,073 in primary care settings, with the large majority attributed to sequencing itself ($4,000). Across cohorts, the study incurred an additional $613 (12% of total WGS costs) per patient randomized to WGS, on average, to confirm monogenic and carrier status variants via Sanger sequencing; and subanalyses of patients randomized to WGS showed an average incremental cost of $111 for each variant reported (p < 0.001). Laboratory personnel time for variant interpretation and reporting increased by 13% for every variant reported (p < 0.001), while overall time demands decreased by an average of 158 min per patient per study year (p = 0.033).
Notably, cardiologists consulted the Genome Resource Center only once during the study, to discuss carrier status findings (compared with 11 consultations in primary care with a variety of questions). In addition, WGS increased the length of cardiology disclosure sessions by an average of 10.7 min (WGS arm, 15.3 min; control arm, 4.6 min; p < 0.001) and primary care disclosure sessions by an average of 18.7 min (WGS arm, 30.1 min; control arm, 11.4 min; p < 0.001). The added length of WGS disclosure sessions increased costs $23 per patient, on average.
Downstream health-care utilization and costs
Health-care utilization and costs from disclosure through 6 months postdisclosure from a health care–sector perspective are summarized in Table 4, and commonly observed procedures during the postdisclosure period are summarized in Supplementary Appendix S9 online. In cardiology care, mean 6-month total downstream costs were $1,560 lower in the WGS arm compared with the control arm (95% confidence interval (CI): –$7,558 to $3,866, p = 0.357), although mean downstream costs in the WGS arm were $700 higher when hospitalizations were omitted (95% CI: –$1,634 to $3,440, p = 0.393). In primary care, mean 6-month total downstream costs were $681 greater in the WGS arm compared with the control arm (95% CI: –$884 to $2,171, p = 0.699) when hospitalizations were included and $374 greater (95% CI: –$991 to $1,570) when hospitalizations were omitted.
Analyses that compared health-care utilization and costs in the postdisclosure period to the predisclosure period showed some increases. Among cardiology participants (Supplementary Appendix S10 online), we observed more imaging tests (all p ≤ 0.047) in both arms in the 6 months following disclosure compared with the 6 months preceding disclosure, although changes over time on costs were observed only for visits (all p ≤ 0.017) and for imaging tests in the WGS arm (p = 0.048). Changes over time in total costs were nonsignificant, with an increase of $2,939 (95% CI: –$2,857 to $8,772) in the control arm and an increase of $1,728 (95% CI: –$3,067 to $6,387) in the WGS arm (Supplementary Appendix S11 online). Changes over time did not differ by randomization status (p = 0.376).
Among primary care participants and comparing the post- with the predisclosure period, we observed more health-care visits in the WGS arm (p = 0.020) and greater visit-related costs in both randomization arms (all p < 0.001). Changes in total costs were nonsignificant, with an increase of $682 (95% CI: –$472 to $1,680) in the control arm and an increase of $1,919 (95% CI: –$1,151 to $2,828) in the WGS arm. Changes over time did not differ by randomization status (p = 0.572).
Subanalyses of participants receiving WGS showed no statistically significant association between mean downstream costs among individuals with a previously unknown monogenic disease risk and individuals without one in either primary care ($4,800 vs. $2,900, respectively, p = 0.76) or cardiology ($6,861 vs. $9,611, respectively, p = 0.47). Analyses on specific categories of costs were also inconclusive (all p ≥ 0.161).
Sensitivity and scenario analyses
Sensitivity analyses (Table 5) compared randomization arms on total costs, including WGS and downstream costs. Varying the WGS costs had the largest impact, where differences between randomization arms were observed at $10,000 but not at $500. Among cardiology patients, differences between randomization arms were observed when the WGS costs were $10,000, when health-care services were assigned cost weights at 50% of CMS fee schedule amounts, and when only immediately attributable services were analyzed. Notably, immediately attributable services (Supplementary Appendix S12 online) were observed for only one cardiology patient in the control arm and one cardiology patient in the WGS arm. Among primary care patients, differences between randomization arms were also observed in all analyses except when WGS costs were $2,500 or less and when health-care services were assigned cost weights at 150% or more of CMS fee schedule amounts (Supplementary Appendix S13 online). Among cardiology patients, omitting secondary findings altogether reduced costs by $69 per patient, whereas among primary care patients, limiting reporting to only pathogenic variants for monogenic disease and carrier status reduced costs by $182 per patient.
Discussion
As the first randomized trial of WGS in primary and specialty care, the MedSeq Project presents a unique opportunity to collect novel data about the costs of WGS. Our data showed that the short-term costs were driven primarily by the costs of sequencing, interpretation, and disclosure, and we did not find evidence that WGS increased downstream health-care costs. Although the MedSeq Project was not powered to draw definitive conclusions about the cost impact of WGS in cardiology and primary care settings, these findings may provide an important early foundation for future trials.
The patient-level data from the MedSeq Project add to the paucity of economic literature surrounding WGS, particularly patient-level data.32 Our findings are in line with previous work that provided microcosting data about whole-exome sequencing,9 and extend them by providing insight about WGS and how costs may vary with different reporting strategies and cost assumptions. Advances in next-generation sequencing approaches are continuing to make WGS less expensive,27 but approximately 25% of the total costs for providing WGS in our study were for steps such as variant confirmation and bioinformatic analyses. Nevertheless, it is possible that the costs associated with these associated steps will also drop, given recommendations to forego confirmation of nucleotide substitutions via Sanger sequencing.33 In addition, the emergence of large genomic data sets such as ExAC,34 improvements in data sharing between laboratories, and innovations in approaches to variant interpretation such as cross-species analyses35 are likely to decrease bioinformatic costs even further by reducing the number of variants that require manual review and classification. Open questions also remain about whether the incremental benefits of WGS are worth its incremental costs compared with approaches such as exome sequencing, which is typically at least 25% less expensive, or gene panels, which can be thousands of dollars less expensive.
Notably, our study showed that WGS had a limited impact on the actions of cardiologists. Cardiologists in our study consulted the Genome Resource Center only once for assistance, spent only 10 additional minutes during disclosure sessions addressing WGS reports, and ordered immediate follow-up testing in response to WGS findings in only one instance. Cardiologists in our study tended to be familiar with genetic testing and regularly reviewed their patients’ family histories, and all of the cardiology patients had received panel genetic testing for their diagnoses prior to disclosure sessions. It is possible that cardiologists had already addressed any important family-history patterns, and that they believed they already had most of the information they needed to judge the importance of WGS findings, despite the novelty of most secondary findings.
While the short-term impact on patient wellbeing was not assessed, WGS did identify health-relevant variants in a large majority of participants, including diagnosis-related variants in half of sequenced cardiology patients and additional monogenic disease risks in 8 cardiology patients and 13 primary care patients. The clinical significance of most of these variants was unclear. Guidelines to help physicians respond to monogenic findings identified in healthy patients or as secondary findings exist for only a few genes and conditions currently, although frameworks defining actionability have been proposed.36 Among our participants, ClinGen Actionability Working Group Evidence-based Summaries existed at that time for 2 of the 8 secondary monogenic disease risks identified in cardiology patients (both in F5), and 5 of the 13 monogenic disease risks identified in primary care patients (HFE (× 2), KCNQ1, TNNT2, and F5).37
Importantly, incorporating WGS into primary care and cardiology settings did not appear to increase downstream health-sector costs over the short term, even among patients who learned about a monogenic risk for disease through secondary findings. We observed more health-care visits in both randomization arms and both cohorts in the 6 months following disclosure relative to the 6 months preceding disclosure, and in cardiology settings, we also observed more imaging and cardiology tests in both arms in the postdisclosure period. These differences are possibly a result of the study design, which mandated an in-person appointment for disclosure that physician participants often also used as well-care visits and health maintenance exams. Regardless, changes in health-care utilization and associated costs did not differ by randomization status. The study may have been too small to detect differences, especially considering the primary analytic approach to include all health-care services and out-of-pocket expenses. In addition, it is also possible that physicians in the MedSeq Project considered few of the WGS findings to be actionable, as evidenced by the small number of immediately attributable services we identified. If a Genome Resource Center had not been available, it is possible that we would have observed additional patient referrals to specialists, particularly among primary care patients who received WGS, and there would have been additional costs associated with these referrals. Nevertheless, the null findings around downstream health costs are notable given that the MedSeq Project’s reporting criteria were far broader than recommendations from the American College of Medical Genetics and Genomics. The MedSeq Project examined a far more extensive list than most research or clinical studies, examining more than 4,600 genes and including findings about carrier status, pharmacogenomic information, and polygenic risk predictions. Our sensitivity analyses suggest that using more liberal reporting criteria doubled the number of potentially health-relevant findings that were reported while increasing overall costs minimally (e.g., less than $200 per patient, on average, to report secondary findings classified as likely pathogenic or uncertain significance: favor pathogenic). Numerous commentators have encouraged strict criteria for the reporting of secondary findings and discourage the use of WGS among healthy populations altogether, citing the possibility that disclosure could initiate follow-up testing and screening that increase costs while accruing limited benefits and possibly causing harms.13,38,39 Our data may begin to assuage those concerns.
Although this was a randomized controlled trial with a planned cost analysis, there are notable limitations. Physicians and patients were enrolled from a single, well-supported health-care system in metro Boston, and physicians and patients in other settings may be less willing to undertake WGS for clinical and financial reasons. It is possible that we would have observed more use of support resources, like the Genome Resource Center, and more follow-up screening and testing if we had enrolled physicians who were less familiar with genomics or had less access to knowledgeable colleagues. As noted above, the sample size was too small to draw conclusions on a population level, and the 6-month time horizon may have been too short to observe the full impact on costs and health benefits. Patient time costs were not assessed, nor were the effects of disclosure on participants’ family members, precluding a complete analysis from a societal perspective. Physicians benefited from extensive study-specific education, and were supported by a Genome Resource Center. Control arm participants received a family-history review that was more rigorous than standard of care, which may have increased their downstream costs. Finally, our patient participants were more educated and less ethnically diverse than the general population. Lack of diversity has been an ongoing problem in genomic research,40 although recent efforts such as the All of Us Research Program have prioritized enrolling more representative populations. Nevertheless, our study provides novel and much-needed data to help decision-makers begin to understand the short-term cost implications of integrating WGS into clinical care, and provides insight about what data are needed to provide more clarity about the economic implications of this technology.
References
McCarthy JJ, McLeod HL & Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med 2013;5:189sr184.
Manolio TA. Incorporating whole-genome sequencing into primary care: falling barriers and next steps. Ann Intern Med 2017;167:204–205.
Armstrong K. Can genomics bend the cost curve? JAMA 2012;307:1031–1032.
Phillips KA, Ann Sakowski J, Trosman J, Douglas MP, Liang S-Y & Neumann P. The economic value of personalized medicine tests: what we know and what we need to know. Genet Med 2014;16:251–257.
Hatz MH, Schremser K & Rogowski WH. Is individualized medicine more cost-effective? A systematic review. PharmacoEconomics 2014;32:443–455.
Diamandis EP. The Hundred Person Wellness Project and Google’s Baseline Study: medical revolution or unnecessary and potentially harmful over-testing? BMC Med 2015;13:5.
Caulfield T, Evans J, McGuire A et al. Reflections on the cost of “low-cost” whole genome sequencing: framing the health policy debate. PLoS Biol 2013;11:e1001699.
Tsiplova K, Zur RM, Marshall CR et al. A microcosting and cost-consequence analysis of clinical genomic testing strategies in autism spectrum disorder. Genet Med 2017;19:1268–1275.
Sabatini LM, Mathews C, Ptak D et al. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the Association for Molecular Pathology. J Mol Diagn 2016;18:319–328.
Sagoo GS, Norbury G, Mohammed S & Kroese M. Whole-Exome Sequencing in Clinical Genetics. A Health Economic Evaluation. PHG Foundation: Cambridge, UK, 2017.
Stark Z, Schofield D, Alam K et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med 2017;19:867–874.
Tan TY, Dillon OJ, Stark Z et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr 2017;171:855–862.
Hegde M, Bale S, Bayrak-Toydemir P et al. Reporting incidental findings in genomic scale clinical sequencing—a clinical laboratory perspective: a report of the Association for Molecular Pathology. J Mol Diagn 2015;17:107–117.
Bennette CS, Gallego CJ, Burke W, Jarvik GP & Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med 2015;17:587–595.
Christensen KD, Roberts JS, Whitehouse PJ et al. Disclosing pleiotropic effects during genetic risk assessment for Alzheimer disease. A randomized trial. Ann Intern Med 2016;164:155–163.
Phillips KA, Pletcher MJ & Ladabaum U. Is the “$1000 genome” really $1000? Understanding the full benefits and costs of genomic sequencing. Technol Health Care 2015;23:373–379.
Christensen KD, Vassy JL, Jamal L et al. Are physicians prepared for whole genome sequencing? A qualitative analysis. Clin Genet 2016;89:228–234.
Vassy JL, Christensen KD, Schonman EF et al. The impact of whole genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med 2017;167:159–169.
Vassy J, Lautenbach D, McLaughlin H et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 2014;15:85.
McLaughlin HM, Ceyhan-Birsoy O, Christensen KD et al. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet 2014;15:134.
Kong SW, Lee IH, Leshchiner I et al. Summarizing polygenic risks for complex diseases in a clinical whole-genome report. Genet Med 2015;17:536–544.
Vassy JL, McLaughlin HL, MacRae CA et al. A one-page summary report of genome sequencing for the healthy adult. Public Health Genomics 2015;18:123–129.
Cirino AL, Lakdawala NK, McDonough B et al. A comparison of whole genome sequencing to multigene panel testing in hypertrophic cardiomyopathy patients. Circ Cardiovasc Genet 2017;10:e001768.
Ramsey SD, Willke RJ, Glick H et al. Cost-effectiveness analysis alongside clinical trials II—an ISPOR Good Research Practices Task Force report. Value Health 2015;18:161–172.
Sanders GD, Neumann PJ, Basu A et al. Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses: second panel on cost-effectiveness in health and medicine. JAMA 2016;316:1093–1103.
Frick KD. Microcosting quantity data collection methods. Med Care 2009;47(7 suppl 1): S76–S81.
National Human Genome Research Institute. The cost of sequencing a human genome. 2016. https://www.genome.gov/sequencingcosts/. Accessed 30 Nov 2016.
Dukhovny D, Dennis CL, Hodnett E et al. Prospective economic evaluation of a peer support intervention for prevention of postpartum depression among high-risk women in Ontario, Canada. Am J Perinatol 2013;30:631–642.
Pagon RA, Tarczy-Hornoch P, Baskin PK et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat 2002;19:501–509.
Hamosh A, Scott AF, Amberger J, Valle D & McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat 2000;15:57–61.
Kalia SS, Adelman K, Bale SJ et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249–255.
Dewey FE, Grove ME, Pan C et al. Clinical interpretation and implications of whole-genome sequencing. JAMA 2014;311:1035–1045.
Baudhuin LM, Lagerstedt SA, Klee EW, Fadra N, Oglesbee D & Ferber MJ. Confirming variants in next-generation sequencing panel testing by Sanger sequencing. J Mol Diagn 2015;17:456–461.
Walsh R, Thomson KL, Ware JS et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192–203.
Chakravorty S & Hegde M. Gene and variant annotation for Mendelian disorders in the era of advanced sequencing technologies. Annu Rev Genomics Hum Genet 2017;18:229–256.
Berg JS, Foreman AK, O’Daniel JM et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med 2016;18:467–475.
ClinGen Clinical Genome Resource. Actionability Working Group Evidence-based Summaries. 2017. https://www.clinicalgenome.org/working-groups/actionability/projects-initiatives/actionability-evidence-based-summaries/. Accessed 17 March 2017.
Yu J-H, Harrell TM, Jamal SM, Tabor HK & Bamshad MJ. Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet 2014;95:77–84.
Kohane IS, Hsing M & Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med 2012;14:399–404.
Bustamante CD, Burchard EG & De La Vega FM. Genomics for the world. Nature 2011;475:163–165.
Acknowledgments
This study was supported by National Institutes of Health grants U01-HG006500, K01-HG009173, KL2-TR001100, and R01-HG007063, and Career Development Award IK2-CX001262 from the US Department of Veterans Affairs Clinical Sciences Research and Development Service. This work was conducted with support from Harvard Catalyst ∣ The Harvard Clinical and Translational Science Center (National Center for Research Resources and National Center for Advancing Translational Sciences, National Institutes of Health grant UL1-TR001102) and financial contributions from Harvard University and its affiliated academic health-care centers. The authors thank 5AM Solutions (Rockville, MD), for their help in customizing the workflow of the “My Family Health Portrait” Web tool for this study. The authors also thank Kenneth Freedberg, Marc Williams, and Ann Wu for their intellectual contributions to the design of these analyses.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Disclosure
The authors declare the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: R.C.G. reports personal fees from Illumina, Helix, GenePeeks, Veritas, and Ohana and is a cofounder with equity in Genome Medical. D.D. reports consulting for Vermont Oxford Network, Gerson Lehrman Group, and ClearView Healthcare Partners and being faculty for Vermont Oxford Network outside the submitted work. The other authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Christensen, K.D., Vassy, J.L., Phillips, K.A. et al. Short-term costs of integrating whole-genome sequencing into primary care and cardiology settings: a pilot randomized trial. Genet Med 20, 1544–1553 (2018). https://doi.org/10.1038/gim.2018.35
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2018.35
Keywords
This article is cited by
-
Behavioral and psychological impact of genome sequencing: a pilot randomized trial of primary care and cardiology patients
npj Genomic Medicine (2021)
-
Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study
Genetics in Medicine (2019)
-
Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy
npj Genomic Medicine (2018)
-
WSG has no impact on short-term healthcare costs
PharmacoEconomics & Outcomes News (2018)