Abstract
Purpose
Nonobstructive azoospermia (NOA) affects 1% of the male population; however, despite state-of-the-art clinical assessment, for most patients the cause is unknown. We capitalized on an analysis of multiplex families in the Middle East to identify highly penetrant genetic causes.
Methods
We used whole-exome sequencing (WES) in 8 consanguineous families and combined newly discovered genes with previously reported ones to create a NOA gene panel, which was used to identify additional variants in 75 unrelated idiopathic NOA subjects and 74 fertile controls.
Results
In five of eight families, we identified rare deleterious recessive variants in CCDC155, NANOS2, SPO11, TEX14, and WNK3 segregating with disease. These genes, which are novel to human NOA, have remarkable testis-specific expression, and murine functional evidence supports roles for them in spermatogenesis. Among 75 unrelated NOA subjects, we identified 4 (~5.3%) with additional recessive variants in these newly discovered genes and 6 with deleterious variants in previously reported NOA genes, yielding an overall genetic etiology for 13.3% subjects versus 0 fertile controls (p = 0.001).
Conclusion
NOA affects millions of men, many of whom remain idiopathic despite extensive laboratory evaluation. The genetic etiology for a substantial fraction of these patients (>50% familial and >10% sporadic) may be discovered by WES at the point of care.
Similar content being viewed by others
Introduction
Infertility, defined as the inability to conceive after 1 year of unprotected intercourse, affects up to 15% of couples worldwide.1,2 Approximately 50% of infertility is male-factor, caused by a wide variety of spermatogenic abnormalities.2 The most severe is nonobstructive azoospermia (NOA), diagnosed by the complete absence of sperm despite lack of obstruction in the reproductive organs3 (Supplementary Table S1 online). NOA accounts for 10 to 15% of male infertility, and thus affects 1% of the adult male population.
The typical clinical evaluation of NOA includes taking a history to rule out cryptorchidism, mumps orchitis, and previous use of chemotherapy or radiotherapy, and conducting a physical examination to rule out obstructive causes that may result in the absence of sperm. Subsequent laboratory investigations include semen analysis, hormonal assessment, chromosomal abnormality assays, and Y-chromosome microdeletions.2,3,4 Together, these analyses explain the cause of only ~30% of cases of NOA.5,6,7 It is generally assumed that many of the remaining idiopathic cases have genetic causes, but although variants in ~55 genes have been associated with NOA in humans8,9 and variants in hundreds of genes are known to impair spermatogenesis in mouse models,10,11,12 there has been limited prospective, point-of-care, next-generation sequencing assessment of males with idiopathic NOA to determine the fraction with genetic causes.13 The challenge in identifying genetic variants in idiopathic males is that the high prevalence of NOA, despite the detrimental effect on reproductive fitness, suggests that high genetic heterogeneity underlies this condition.5,14,15 Thus, in outbred populations, large numbers of affected males would need to be recruited to find the few who may share the same causative mutations, or rare, damaging mutations disrupting key genes along the delicate, evolutionarily conserved spermatogenesis pathway.11,14 Conversely, in the setting of consanguinity, novel causative NOA genes may be discovered from single families using next-generation sequencing technologies.16,17,18
Based on these considerations, we hypothesized that whole-exome sequencing (WES) could be applied to identification of novel genes/variants associated with idiopathic NOA in endogamous populations. Our approach had two parts. First, we investigated new genetic causes of NOA by evaluating multiplex families with idiopathic brothers from the Middle East, where the cultural practice of consanguinity and traditionally large families ensure the availability of multiple affected as well as fertile siblings. We assessed 8 such families by comparing their WES data with exome sequencing data on 1,376 population-matched controls to identify rare, putatively deleterious recessive variants that segregate with affected siblings, affect spermatogenesis genes, and may explain the NOA phenotype.19 In the second part, when this analysis identified 5 novel candidate genes, we combined this information with that for 57 genes in which protein-altering variants had been previously linked to NOA from the literature to create a 62-gene NOA gene panel, and investigated the utility of this panel as a point-of-care screening tool for NOA cases thought to be idiopathic. Interestingly, WES assessment of a cohort of 75 idiopathic NOA showed that 10 subjects (13.3%) harbored additional rare, recessive, predicted damaging variants in the NOA gene panel, suggesting that many idiopathic cases may have a genetic etiology for which WES investigation would be well suited.
Materials and methods
Study subjects
The study was performed with the approval of Institutional Review Boards for Human Subjects Research at Hamad Medical Corporation Hospital and Weill Cornell Medicine–Qatar. Two cohorts with NOA were assessed: A—familial, used for identification of novel NOA-associated genes, and B—sporadic, used to test the value of a point-of-care NOA gene panel and WES to assess idiopathic NOA subjects for a possible genetic basis. Affected subjects were young and fit, with no comorbidities or health issues. All subjects gave informed consent and underwent thorough history taking, physical examination, and semen analysis, which was done according to the World Health Organization’s 2010 recommendations to rule in azoospermia. Supplementary Figure S1 online is a schematic of the overall study strategy. Semen samples were examined after being centrifuged at 3,000g for 15 min. A hormonal profile was recorded, including follicle-stimulating hormone (normal range 1–19 IU/L), luteinizing hormone (1–9 IU/L), prolactin (73–407 mIU/L), and total testosterone (10.4–35 nmol/L) and estradiol (n = 73–275 pmol/L). All hormonal levels were recorded at the initial visit, i.e., prior to any hormonal treatment for infertility.
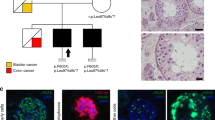
Infertile subjects recruited to this study had NOA based on standard criteria20 (Supplementary Table S1 online), including being negative for evidence of obstructive azoospermia, Y-chromosome microdeletions, or karyotyping abnormalities (see Supplementary Methods online). None of the patients were receiving chemotherapy or radiotherapy or had a history of cryptorchidism. For the familial cohort (A), we required two affected brothers as well as at least one proven fertile male in the family (living father and/or at least one brother with children). A total of 37 individuals in eight families were assessed (Table 1,Figure 1, Supplementary Table S2 online). For the sporadic cohort (B), 75 individuals with idiopathic NOA were evaluated (Supplementary Table S3 online) and compared with 74 men with a history of fathering at least 1 child (Supplementary Table S4 online).
Eight consanguineous families with nonobstructive azoospermia (NOA). For each family, whole-exome sequencing was used to assess both affected siblings (black shaded boxes) with NOA plus at least one fertile male per family (the father and/or additional sibling(s) with children—denoted “with children” where applicable). A total of 37 individuals were sequenced (thick black borders), other family members appear in gray. Numbers inside boxes indicate number of siblings of a given sex (square, brothers; circle, sisters). For five families, candidate pathogenic variants explaining infertility were discovered. These are indicated in Human Genome Variation Society format for each family, and the segregation of the variant is shown under each sequenced individual, wild-type allele, variant allele; Y, signifying the variation is on the X chromosome in a male.
WES and analysis pipeline
A total of 186 individuals underwent WES for this study: 37 from families and 149 from the idiopathic cohort. Blood-extracted DNA was exome-captured using Agilent’s SureSelect v5 platform (Santa Clara, CA) and sequenced on Illumina’s HiSeq 2000 platform (San Diego, CA). Reads were mapped to the human reference genome GRCh37/hg19 using BWA 0.5.9 (https://bio-bwa.sourceforge.net/). Average exome depth of coverage was 94.6 × (Supplementary Tables S5–S7 online). Variants were called using SAMtools 0.1.17, annotated using SeattleSeq v138 (ref.21) and filtered to identify Mendelian variants. Briefly, variants were removed if (i) SAMtools quality score was <100 (http://samtools.sourceforge.net/); (ii) segregation did not fit the pedigree; (iii) minor allele frequency was >1% in any database, including the Genome Aggregation Database (gnomAD), containing 123,136 exomes (previously, Exome Aggregation Consortium (ExAC)) and 15,496 genomes;22 (iv) the variant affected: 3′ or 5′ untranslated regions, noncoding exons, or intronic sequences except canonical splice sites; and (v) the variant was not predicted damaging by PolyPhen23 and CADD24 or was not predicted to alter a high-conserved residue by PhastCONS25 and GERP.26
Assessment of variant rarity benefited from our allele-frequency data on 1,376 ethnically matched individuals who had undergone WES to study the Qatari genome.19 For each variant passing the stringent filtration criteria, literature and gene-expression databases were searched for association to infertility or spermatogenesis. All evidence was manually curated and validated (see Supplementary Methods online), and genes linked to infertility were identified accordingly. To confirm that candidate recessive variants occurred in runs of homozygosity, the homozygosity heterogeneous hidden Markov model tool (H3M2) was run with default parameters on all sequenced family members to produce runs of homozygosity plots per chromosome, as described by Magi et al.27
Tissue-specific expression and enrichment of candidate genes in testis
All genes harboring candidate pathogenic variants from WES curation were evaluated for likelihood of being involved in disease by examining data from the Genotype-Tissue Expression Project (http://www.gtexportal.org), which contains RNA expression data for all genes in 32 human tissues,28 and from the Human Protein Atlas (http://www.proteinatlas.org), which contains tissue-specific protein expression data.29 Expression levels of 12 candidate genes identified in the familial cohort were plotted using the R heatmap() (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/heatmap.html) function showing the lowest (green) to highest (red) expression levels in 32 tissues. Five genes with highly tissue-specific expression were replotted in three dimensions to demonstrate highly specific tissue expression in testis. Only 1,051 of 19,556 genes in the Human Protein Atlas had similarly high testis-specific expression (at least fivefold higher expression in testis over second highest tissue). Statistical enrichment was assessed using a chi-squared test.
Point-of-care assessment of NOA genes in unrelated subjects with idiopathic NOA
A separate cohort of 75 unrelated subjects with idiopathic NOA and 74 unrelated ethnically matched fertile men was evaluated by WES to detect additional recessive, high-quality, protein-altering, predicted damaging variants in the familial genes (Supplementary Table S8 online). This same cohort was evaluated for additional possibly causative variants by querying the exome data for variants in known NOA genes. This NOA gene panel was created via detailed evaluation of the literature for genes with protein-coding pathogenic variants (or exonic deletions) associated with human NOA, by searching for the terms: nonobstructive azoospermia, Sertoli cell only, maturation arrest, hypospermatogenesis, and spermatogenic failure. Genes related to other forms of infertility (e.g., obstructive azoospermia, motility defects, oligospermia, or hormonal infertility), genes reported without specific mutations (e.g., through RNA or expression studies), genes in genomic duplications, and genes implicated by proximity to significantly associated genome-wide association study single-nucleotide polymorphisms were excluded. This analysis led to identification of 57 genes in the following four categories: (i) genes in critical AZF regions on the Y chromosome (n = 9); (ii) candidate genes found to harbor rare coding mutations including exonic deletions in NOA cohorts (“candidate genes—causative,” n = 13); (iii) genes in which protein-altering variants have been statistically associated with NOA through large case-control studies, but some of which may appear at lower frequency in controls (“candidate genes—associated,” n = 29); and (iv) genes identified in familial or linkage studies segregating with NOA (“identified in families,” n = 6). Variants in this panel, and in candidate genes identified in the family, were evaluated in the same cohort of 75 idiopathic NOA subjects and 74 fertile men, focusing on recessive, high-quality, protein-altering, predicted damaging variants. The significance of observed versus expected mutations in cases versus controls was assessed using Fisher’s exact test.
Results
Identification of NOA-related genes in the families
All available members from eight multiplex families (Figure 1) were sequenced to a mean depth of 99.5 ×, with the average subject having 99.1% and 92.3% of target exome bases covered by at least 4 and 20 unique reads, respectively (Supplementary Table S5 online). All samples underwent individual as well as joint (familial) variant calling and annotation. Candidate variants were identified as being recessive, segregating appropriately within the family (not homozygous in father and fertile male siblings), protein-altering, affecting evolutionarily conserved residues and predicted damaging by a range of prediction tools (see Materials and Methods, Supplementary Figure S1 online). Additionally, variant frequency was assessed to eliminate common variants (minor allele frequency >1%) present in four public variation databases (i.e., dbSNP, gnomAD, NHLBI GO-ESP, and 1000 Genomes), as well as against a database of 1,376 Qatari exomes19 to eliminate population-specific polymorphisms. This process dramatically reduced the total number of candidate exome variants per family from >20,000 to a median of 2 per family (range: 0–4, see Supplementary Table S9 online). In total, only 12 genes harboring putatively deleterious variants remained in six of the eight families (see Supplementary Table S9 online).
To investigate whether any of these 12 genes may be related to NOA, we first examined the tissue expression patterns for each gene, obtained from the Human Protein Atlas (http://www.proteinatlas.org). Surprisingly, global expression evaluation in 32 tissues demonstrated that 5 of the 12 genes (CCDC155, NANOS2, SPO11, TEX14, and WNK3) had remarkably high testis-specific expression, one in each of five families (Figure 2a). We evaluated this further, both visually (Figure 2b) and by calculating the ratio of testis expression to the second highest site of expression for each gene (Table 2 and Supplementary Figure S2 online). On average, the five genes had 60-fold higher expression (range: 6.3- to 120-fold) in the testis over the second highest expression site. By comparison, of 19,557 genes in the protein atlas, 3,529 genes have their highest relative expression levels in testis. Of these, only 1,051 had testis-specific expression (at least fivefold higher expression in the testis over the second-highest ranking tissue site), representing a significant enrichment in our data set of testis-specific genes (chi-squared p = 8.49 × 10−7). This enrichment of rare homozygous damaging mutations in testis-specific genes, and the distribution of these five genes (each in a different NOA family), suggested these genes are candidates for causing NOA in five families (Table 1, additional pathogenicity scores and details in Supplementary Table S10 online). As expected, all autosomal candidate variants were observed in long runs of autozygosity, consistent with their being recessive founder alleles homozygous due to consanguinity (Supplementary Figure S3 online).
Expression patterns of genes harboring putatively damaging variants in nonobstructive azoospermia (NOA) families. (a) Gene expression heat map of all 12 genes harboring rare, predicted damaging recessive variants in NOA families. For each family, expression patterns for all genes surviving stringent filtration were retrieved from the Human Protein Atlas and plotted in a heat map as described in Materials and Methods. Six families had such variants (Supplementary Table S2 online), revealing five candidate genes with testis-specific expression (in bold type and replotted in (b) to demonstrate tissue-specificity). *Additional variants in this gene found in the idiopathic NOA cohort (Table 3); +Mouse model also infertile due to abnormal spermatogenesis (Table 2). RKPM, reads per kilobase per million. (b) Five candidate infertility genes with highly specific testis expression. Normalized expression data obtained from the Human Protein Atlas (http://www.proteinatlas.org; see Materials and Methods for details)29 is plotted for the five candidate genes of interest across a wide range of tissues, sorted first by increased testis expression levels and then by increased expression in other tissues (additional details in Supplementary Figure S2 online).
To complement expression analysis, we also investigated the reported roles of these five genes in the literature or mouse databases (see Materials and Methods). For four of the five genes (Ccdc155, Nanos2, Spo11, and Tex14) mouse knockout impairs spermatogenesis, while for Wnk3, knockout of a gene it activates (Nkcc1) leads to male mouse infertility. Remarkably, reported histological phenotypes from mouse testes for each gene were consistent with histological phenotypes in the affected brothers, where available (Table 2; see Discussion).
Point-of-care assessment of a sporadic NOA cohort
As a follow-up to the familial cohort, we sequenced the exomes of 75 unrelated individuals with idiopathic NOA recruited at the point of care, to assess the presence of additional mutations in candidate familial genes. For controls, we sequenced the exomes of 74 ethnically matched, confirmed fertile men (Supplementary Tables S3,S4,S7, and S7 online). Overall, both cohorts had a similar burden of high-quality variants across different genic categories (Supplementary Table S8), consistent with their being ethnically matched and sequenced to similarly high depths on the same platform (see Materials and Methods, Supplementary Tables S6 and S7). We again implemented stringent filtration criteria (allele frequency, impact, conservation scores) to discover high-quality recessive, deleterious variants in these groups. Of the 12 candidate genes identified in the familial cohort, we found additional evidence for 3 genes, in four subjects (Table 3). Notably, these three genes (NANOS2, TEX14, and WNK3) were among the five testis-specific familial genes, further supporting their role in pathogenesis among the 12 possible candidates (Figure 2). No similarly deleterious variants in these 12 genes were found in the fertile controls.
Assessment of a sporadic cohort for additional variants in reported NOA genes
Given the genetic heterogeneity of NOA, we leveraged the WES data from the sporadic cohort and controls to search for additional variants likely to cause idiopathic NOA. A detailed literature search identified 57 genes in which protein-altering variants were associated with NOA (Supplementary Table S11 online; see Materials and Methods for details on panel curation).
Within the group of 75 idiopathic NOA subjects, 6 individuals (7.5%) harbored deleterious recessive variants in panel genes (Table 3 and Supplementary Table S8 online). None of these specific variants had previously been described in NOA in humans, consistent with their putatively detrimental effect on reproductive fitness and the high allelic heterogeneity expected of this disease. By comparison, no similarly deleterious variants in these 57 genes were observed in the fertile controls (Supplementary Table S8 online).
Altogether, recessive variants in the previously reported and newly discovered familial genes were present in 10 of 75 (13.3%) idiopathic subjects (Table 3). Five of these were putative loss-of-function variants (two affecting splice sites, two resulting in frameshifts, and one start-loss), and the remaining five were all predicted highly damaging missense variants (median PolyPhen score, 0.98; CADD, 18.5; and GERP, 3.9). In contrast to the idiopathic NOA group, none of the 74 fertile males had recessive deleterious variants in any of these genes (Fisher’s p = 0.001; Table 3 and Supplementary Table S8 online). This finding supports the utility of combining a NOA gene panel (Supplementary Table S12 online) and WES to evaluate idiopathic NOA at the point of care.
Discussion
Despite the high prevalence of male infertility, discovery of new causative genes has remained challenging in human cohorts. This may be partially explained by the high genetic heterogeneity expected based on model organism studies, and by the sporadic nature of the condition in outbred populations; in this setting, probably causative variants would be indistinguishable from private mutations, making their interpretation as causal challenging.6,8,9,14,15 In the present study, we overcame these challenges by focusing on recessive variants in the setting of high consanguinity, where homozygosity for rare, predicted damaging alleles in affected siblings, absent from fertile males in the family, provides initial genetic evidence for involvement in disease. We combine this evidence with observation of remarkable testis-specific expression, mouse model phenotypes where available, and additional investigation in a sporadic idiopathic cohort to support the identification of five candidate human NOA genes.
Identification of novel human NOA genes
Combining the genetic evidence from the familial and idiopathic NOA cohorts, the specificity of expression in human testes, and data from mouse studies, we obtained evidence for five candidate genes for human NOA (TEX14, SPO11, NANOS2, WNK3, and CCDC155).
Mouse Tex14 is a testis-expressed protein that localizes to intercellular bridges, where it recruits bridge components to coordinate cytokinesis in the first meiotic division. When knocked out, male mice are sterile due to maturation arrest (MA) and increased germ cell apoptosis.30 Both brothers in the first family (NOA001) with the TEX14 variant (p.Arg85Leu) were diagnosed with MA, whereas the two additional individuals (I042 and I060, Table 3) from the unrelated cohort (with homozygous frameshift and splice-site variants, p.Ser1255fs and c.555-5 T>G, respectively) were diagnosed with Sertoli cell only (SCO).
SPO11 encodes a type II–like topoisomerase expressed in meiosis, where it introduces double-strand DNA breaks that are critical for synaptonemal complex formation between homologous chromosomes during recombination. When knocked out in mice, meiotic chromosomes fail to synapse and spermatocytes do not progress beyond the zygotene stage. Consistent with this biological function, both brothers in the second family (NOA002) with the p.Glu186Lys variant had a histopathology diagnosis of MA. No additional unrelated individuals were found with variants in this gene.
NANOS2 encodes an RNA-binding protein whose knockout leads to male-specific, complete germ cell loss in both Drosophila and mouse.31 Both of the brothers in family 4 with the missense variant (p.Gly70Arg) had SCO; the additional sporadic subject (I074, Table 3), in whom a homozygous start-loss variant (p.Met1?) was discovered, presented with MA.
CCDC155 (mouse Kash5) encodes a dynein-binding protein on the outer nuclear membrane that connects Sun1-bound telomeres to the cellular microtubule complex to enable movement of sister chromatids and alignment of homologues during meiosis. Knockout of Kash5 in male mice leads to meiotic arrest, similar to Tex14 and Spo11, matching the histological finding of spermatocyte MA observed in the brothers in the family NOA008, who share a predicted deleterious variant p.Leu535Gln.
Finally, the relationship between WNK3 and NOA remains the most challenging to establish. Functionally, WNK3 belongs to the WNK (with no lysine (K)) serine–threonine kinases family that regulate cation–chloride transporters in multiple tissues.32 WNK3 is a potent activator of NKCC1, and targeted disruption of Nkcc1 in mouse, which is also natively expressed in Sertoli cells, leads to defects in spermatogenesis and complete absence of spermatocytes.33 This phenotype is consistent with the idiopathic NOA subject (I030, Table 3) who presented with SCO, and in whom we found a predicted damaging variant (p.Glu913Lys) in WNK3, while no histology was available for the affected brothers from family NOA006 to investigate phenotypic concordance. However, murine Wnk3 appears dispensable for fertility, as one study generated Wnk3−/− females from Wnk3Y/− fathers,34 suggesting that the link between this gene and infertility may not be identical across mouse and man. In that report, for example, the direct effect of Wnk3 deletion on spermatogenesis was not investigated, and thus the deletion may cause subtle spermatogenic impairment but not complete loss of fertility in mice. Further, the role of the two orthologs may not be identical in the two species; for example, while WNK3 expression is highly enriched in testis in humans (Figure 2), mouse Wnk3 expression is equally high in the brain, kidney, thymus, and stomach, and moderately expressed in fetal liver, lung, heart, and ileum.34 Additionally, there is evidence in mice that the Wnk1/Spak axis compensates for Wnk3 function in vivo 35 and that both Wnk1 and Wnk4 may compensate for Wnk3 activity,32 possibly explaining murine tolerance to loss of Wnk3. In contrast, the other human WNK family members are expressed only moderately in testes, and WNK3 appears to be critical for human biology: human WNK3 is among the top 5% most constrained genes in the human genome ExAC missense constraint, z = 1.57) and is considered extremely intolerant to loss-of-function variants (ExAC probability of loss-of-function intolerance, pLi = 1.0).22 This evidence, combined with the testis-specific expression and observation of deleterious mutations in 3 of 85 azoospermic men in our cohort, makes it a strong candidate for human infertility. However, additional functional work may be required to elucidate mechanisms and reconcile the mouse and human data; e.g., it is possible that WNK family members do not adequately compensate for loss of function in humans or that point mutations in this gene act in a gain-of-function or dominant negative manner to disrupt some biological pathway, whereas a total gene deletion is tolerated due to paralog compensation.
The utility of WES in the assessment of idiopathic NOA at the point of care
To assess the usefulness of WES in point-of-care evaluation of idiopathic NOA, we assessed individuals for pathogenic variants in newly discovered genes as well as known NOA genes by combining them on a NOA gene panel (Supplementary Table S12 online). This analysis revealed that 10 (13.3%) of the NOA subjects considered idiopathic, but none of the fertile controls, harbored rare recessive, putatively damaging variants in genes already linked to human NOA. Four of the subjects had mutations in three of the genes discovered in our familial cohort (TEX14, NANOS2, and WNK3), providing further evidence for their contribution to NOA.
This study demonstrates that NOA patients who remain idiopathic following routine investigation may benefit from WES with a reasonable likelihood of finding a candidate variant, i.e., a rare or novel coding variant altering a highly conserved amino acid in a gene or pathway that has previously been shown to affect spermatogenesis. While this study benefits from the enrichment for homozygosity due to consanguinity, studies in outbred populations may similarly focus on discovering variants disrupting candidate panel genes that likely impair spermatogenesis. This approach could detect rare variants underlying common disease architecture, similar to those observed in several other highly prevalent sporadic diseases affecting millions around the world, such as hypertension or hypertriglyceridemia.36,37 If large, outbred case-control cohorts are being investigated instead of a highly consanguineous population, our NOA gene panel (Supplementary Table S12 online) may be expanded to include genes implicated from association studies, genes in copy-number duplications, and genes impairing spermatogenesis in other model organisms. However, each of these categories would impose additional challenges in interpretation and would require larger cohorts to provide sufficient power based on genetic model; i.e., our ability to recruit a highly consanguineous population significantly increases the power of finding rare, recessive, protein-altering variants with large effects, even in a modest-sized sporadic cohort. For example, in a recent study investigating a panel of 107 genes in 1,112 NOA subjects, diagnostic variants were found in only 1.5% of subjects.13 The discrepancy between our two rates could be due to the difficulty of finding recessive variants in outbred patients, as compared with the naturally much higher rate in consanguineous populations.
Identification of genetic variants is useful for NOA patients on several levels. First, it is possible that understanding the biological defect underlying this disease could lead to personalized treatments targeting affected pathways. Second, genetic stratification of patients may improve the a priori assessment of the likelihood of finding spermatocytes if the patient agrees to undergo surgical retrieval procedures. For example, one recent study in idiopathic familial NOA reported a significantly lower success rate of surgical sperm retrieval compared with nonfamilial idiopathic cases.7 Third, knowing the causative variant in each patient may enable the screening of prospective in vitro fertilization embryos to ensure pathogenic infertility variants are not transmitted to future male children.
Conclusion
In summary, the study of human infertility may benefit from the introduction of WES in the clinical workup for all patients who remain idiopathic after routine evaluation. When combined with functional evidence implicating hundreds of genes in murine reproductive abnormalities12 and testis-specific gene expression patterns in humans, this strategy may lead to a substantial diagnostic rate and discovery of novel male infertility genes, with implications for future treatments enhancing fertility or the development of molecular contraceptives.
References
Thonneau P & Spira A. Prevalence of infertility: international data and problems of measurement. Eur J Obstet Gynecol Reprod Biol 1991;38:43–52.
Agarwal A, Mulgund A, Hamada A & Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol 2015;13:37.
Cooper TG, Noonan E, von Eckardstein S et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update 2010;16:231–245.
Sharlip ID, Jarow JP, Belker AM et al. Best practice policies for male infertility. Fertil Steril 2002;77:873–882.
Hwang K, Yatsenko AN, Jorgez CJ et al. Mendelian genetics of male infertility. Ann NY Acad Sci 2010;1214:E1–E17.
Lee JY, Dada R, Sabanegh E, Carpi A & Agarwal A. Role of genetics in azoospermia. Urology 2011;77:598–601.
Arafa MM, ElBardisi HT, AlSaid SS et al. Outcome of microsurgical testicular sperm extraction in familial idiopathic nonobstructive azoospermia. Andrologia 2015;47:1062–1067.
Zorrilla M & Yatsenko AN. The genetics of infertility: current status of the field. Curr Genet Med Rep 2013;1:247–260.
Lin YN & Matzuk MM. Genetics of male fertility. Methods Mol Biol 2014;1154:25–37.
Matzuk MM & Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008;14:1197–1213.
Iguchi N, Tobias JW & Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci USA 2006;103:7712–7717.
Yatsenko AN, Iwamori N, Iwamori T & Matzuk MM. The power of mouse genetics to study spermatogenesis. J Androl 2010;31:34–44.
Oud MS, Ramos L, O’Bryan MK et al. Validation and application of a novel integrated genetic screening method to a cohort of 1,112 men with idiopathic azoospermia or severe oligozoospermia. Hum Mutat 2017;38:1592–1605.
Aston KI & Conrad DF. A review of genome-wide approaches to study the genetic basis for spermatogenic defects. Methods Mol Biol 2013;927:397–410.
Seminara SB, Crowley WF Jr. Genetic approaches to unraveling reproductive disorders: examples of bedside to bench research in the genomic era. Endocr Rev 2002;23:382–392.
Ayhan O, Balkan M, Guven A et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet 2014;51:239–244.
Okutman O, Muller J, Baert Y et al. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 2015;24:5581–5588.
Ramasamy R, Bakircioglu ME, Cengiz C et al. Whole-exome sequencing identifies novel homozygous mutation in NPAS2 in family with nonobstructive azoospermia. Fertil Steril 2015;104:286–291.
Fakhro KA, Staudt MR, Ramstetter MD et al. The Qatar genome: a population-specific tool for precision medicine in the Middle East. Hum Genome Var 2016;3:16016.
World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 2010. http://www.who.int/reproductivehealth/publications/infertility/9789241547789/en/. Accessed 8 November 2017.
Ng SB, Turner EH, Robertson PD et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272–276.
Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291.
Adzhubei IA, Schmidt S, Peshkin L et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM & Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315.
Siepel A, Bejerano G, Pedersen JS et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005;15:1034–1050.
Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S & Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005;15:901–913.
Magi A, Tattini L, Palombo F et al. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics 2014;30:2852–2859.
GTex Consortium. The Genotype-Tissue Expression (GTEx) Project. Nat Genet 2013;45:580–585.
Uhlen M, Fagerberg L, Hallstrom BM et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419.
Greenbaum MP, Yan W, Wu MH et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci USA 2006;103:4982–4987.
Tsuda M, Sasaoka Y, Kiso M et al. Conserved role of nanos proteins in germ cell development. Science 2003;301:1239–1241.
Rodan AR & Jenny A. WNK kinases in development and disease. Curr Top Dev Biol 2017;123:1–47.
Pace AJ, Lee E, Athirakul K, Coffman TM, O’Brien DA & Koller BH. Failure of spermatogenesis in mouse lines deficient in the Na(+)-K(+)-2Cl(-) cotransporter. J Clin Invest 2000;105:441–450.
Begum G, Yuan H, Kahle KT et al. Inhibition of WNK3 kinase signaling reduces brain damage and accelerates neurological recovery after stroke. Stroke 2015;46:1956–1965.
Mederle K, Mutig K, Paliege A et al. Loss of WNK3 is compensated for by the WNK1/SPAK axis in the kidney of the mouse. Am J Physiol Renal Physiol 2013;304:F1198–1209.
Ji W, Foo JN, O’Roak BJ et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet 2008;40:592–599.
Romeo S, Yin W, Kozlitina J et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest 2009;119:70–79.
Romanienko PJ & Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000;6:975–987.
Suzuki A & Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev 2008;22:430–435.
Horn HF, Kim DI, Wright GD et al. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol 2013;202:1023–1039.
Acknowledgments
These studies were supported, in part, by the Qatar Foundation and the Weill Cornell Medical College in Qatar and NPRP 5-436-3-116. We thank K. Ahmed, S. Mahadik, F. Al-Maadid, H. Zeriri, and T. Odeh for help with this project, and N. Mohamed for help in preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Fakhro, K.A., Elbardisi, H., Arafa, M. et al. Point-of-care whole-exome sequencing of idiopathic male infertility. Genet Med 20, 1365–1373 (2018). https://doi.org/10.1038/gim.2018.10
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2018.10
Keywords
This article is cited by
-
Low androgen signaling rescues genome integrity with innate immune response by reducing fertility in humans
Cell Death & Disease (2024)
-
A novel homozygote nonsense variant of MSH4 leads to primary ovarian insufficiency and non-obstructive azoospermia
Molecular Biology Reports (2024)
-
Burden of Mendelian disorders in a large Middle Eastern biobank
Genome Medicine (2024)
-
Stepwise mini-incision microdissection testicular sperm extraction in NOA patients with a history of cryptorchidism: a case–control study
Basic and Clinical Andrology (2023)
-
Sertoli cell-only syndrome: advances, challenges, and perspectives in genetics and mechanisms
Cellular and Molecular Life Sciences (2023)