Abstract
Purpose
The advent of next-generation sequencing resulted in substantial increases in the number of variants detected, interpreted, and reported by molecular genetics diagnostic laboratories. Recent publications have provided standards for the interpretation of sequence variants, but there are currently no standards regarding reinterpretation of these variants. Recognizing that significant changes in variant classification may occur over time, many genetics diagnostic laboratories have independently developed practices for variant reinterpretation. The purpose of this study is to describe our laboratory approach to variant reinterpretation.
Methods
We surveyed eight genetics diagnostic laboratories in Canada and the United States.
Results
Each laboratory had differing protocols, but most felt that clinically relevant changes to variant classifications should be communicated to ordering providers. Based on results of this survey and our experience, we developed a cost-effective and resource-efficient approach to variant reinterpretation.
Conclusion
Ongoing variant reinterpretation is required to maintain the highest standards for delivering genetics laboratory services. Our approach to variant reinterpretation offers an efficient solution that does not compromise accuracy or timely delivery of genetics laboratory services.
Similar content being viewed by others
Introduction
The advent of next-generation sequencing has allowed the simultaneous analysis of numerous disease genes by molecular genetics diagnostic laboratories. This has resulted in a substantial increase in the number of sequence variants detected, interpreted, and reported. Recent publications have provided guidelines for the standardization of terminology and classification of these variants.1, 2 While these guidelines have provided much-needed direction for variant interpretation, there remains an absence of clear and specific guidelines regarding if and/or how often these variants should be reinterpreted, and, if the variant classification changes over time, how that should be communicated to health-care providers. Several publications have emphasized an urgent need for clarity regarding the roles and responsibilities of laboratory staff, as well as primary and genetics health-care providers in how to manage the increased amount of genetic information and evolving nature of variant classification.3, 4, 5
In a joint consensus paper, the American College of Medical Genetics and Genomics and the Association for Molecular Pathology recommended that laboratories develop clear policies regarding their variant reinterpretation workflows, and communicate these policies to referring providers.1 Additionally, laboratories were encouraged to suggest that referring providers contact them if updated information is desired regarding a variant of uncertain clinical significance. Finally, they stated that if a laboratory reclassifies a variant that was initially reported as pathogenic or benign, the laboratory should consider proactively amending any previously issued reports impacted by this change.1 The issue of variant reinterpretation was briefly discussed in the EuroGentest guidelines from the European Society of Human Genetics,6 which stated that laboratories should not be expected to systematically review all previously reported variants. Also, if laboratory personnel were made aware of a change in variant classification, it was advised to communicate with the referring provider(s) and amend all previously issued reports impacted by the change.
These publications offer a basic framework for variant reinterpretation. While specific guidelines were not provided, both consensus articles1, 6 suggested that if variant reinterpretation is desired, referring providers should request this information from the laboratory. However, if a laboratory becomes aware of a potentially clinically significant change to the information previously reported, both publications agree that the laboratory should consider communicating this information to the referring providers. Carrieri et al.5 described the attitudes of genetic health-care providers and laboratory scientists in the United Kingdom regarding recontacting former patients with updated genetic information; their participants emphasized the varied and conflicting views on this issue. Most participants agreed in principle that recontacting patients when clinically indicated would be ideal, but expressed concerns about the resources required for this work, the lack of infrastructure to support it, and the lack of guidelines in this area.
Our laboratory has abided by the general principles of variant reinterpretation outlined above for many years. However, with an increase in variants reported due to expansion of our genetics laboratory services to include next-generation sequencing panels in 2015, challenging scenarios regarding variant reinterpretation have arisen. These scenarios have highlighted the lack of published recommendations in this area, and illustrated the need for evidence-based guidelines. As there are many stakeholders impacted by variant reinterpretation, and the implications of these policies are far-reaching, the development of standardized guidelines is an essential next step. Important considerations include whether the laboratory should withhold or communicate potentially actionable medical information to a referring provider that was not explicitly requested, the logistical barriers to communication between the laboratory and a referring provider or a provider and a patient, as well as increased variant reinterpretation and communication workload for laboratory staff. Additionally, laboratories must have the infrastructure to easily identify all patients found to have a specific variant of interest; without such capability, it would not be feasible for a laboratory to contact referring providers of previous patients regarding a variant classification change. The purpose of this study was to expand on the framework provided by previous publications1, 6 and to describe the standardized approach to variant reinterpretation developed by our laboratory.
Materials and methods
Identifying evidence for guideline development
After identifying the lack of specific guidelines for variant reinterpretation, we contacted other genetics diagnostic laboratories to capture their insights and individual practices. We spoke with a representative from each of eight genetics laboratories in Canada and the United States that routinely perform sequence variant interpretation; see Supplementary Table S1 online for additional information about the laboratories surveyed. These telephone conversations included the following questions: (i) does your laboratory have a protocol for systematic review of the clinical significance of all previously identified variants?; (ii) does your laboratory accept requests to reanalyze the clinical significance of a previously reported variant from the referring provider?; (iii) if your laboratory identifies a previously reported variant in another unrelated proband, does your laboratory reinterpret this variant?; and (iv) if upon variant reinterpretation the variant classification changes, does your laboratory contact the referring provider of previous patients found to have this variant?
In addition to conducting this survey, we further discussed the complexities of variant reinterpretation with laboratory staff representing nine institutions in Ontario at the 2016 Ontario Genetics Secretariat Planning Retreat meeting, as well as with colleagues from five institutions across Canada via e-mail discussion. Finally, we have also discussed this issue with clinical geneticists and genetic counselors at our own institution.
Results
Survey of genetics diagnostic laboratories
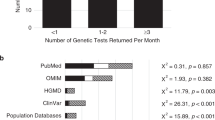
In response to question 1, none of the eight laboratories surveyed stated that they systematically review significance of previously analyzed variants. Two of the laboratories have variant review teams that perform variant reinterpretation if they discover that additional information about a specific variant is published in the literature, but they do not proactively search for additional information available regarding all variants.
In response to question 2, all laboratories surveyed indicated that they will perform variant reinterpretation if requested, at no charge to the referring provider or patient. Additionally, in response to question 3, all laboratories have a policy to perform a variant reinterpretation if it is detected in another proband, and significant time has elapsed since the previous interpretation. The policy for the amount of time elapsed varied amongst laboratories; while some laboratories did not have a set time period, most perform variant reinterpretation if either more than 6 or 12 months have elapsed since the previous variant analysis.
Question 4 (If upon variant reinterpretation, the variant classification changes, does your laboratory contact the referring provider of previous patients found to have this variant?) prompted the most discussion during each telephone conversation. All laboratory representatives agreed that this is a challenging scenario with many considerations, and that clear guidelines for how to address this would be welcomed. One laboratory indicated that they amend all previous reports if a variant classification changes, regardless of the type of change. If the classification change is potentially clinically significant (e.g., change from a variant of uncertain significance to likely pathogenic or pathogenic classification, change from likely pathogenic to likely benign), seven of the eight laboratories indicated that they will attempt to contact the referring provider for all patients found to have the variant to discuss the change, and will provide an amended report if requested. Anecdotally, several laboratory representatives reported that referring providers have been appreciative of receiving this type of communication, but that on occasion they have heard concern from the provider that they may not be able to contact the patient, particularly if significant time has passed since the original test was ordered.
Additional personal communication
E-mail discussions with genetics diagnostic laboratory directors across Canada revealed that most laboratories do not have set protocols for variant reinterpretation. While some laboratories treat each scenario involving a change in variant classification on a case-by-case basis, most laboratory staff agreed that if there is clinically significant change in classification, this information should be communicated to the referring provider, assuming adequate resources are available. Additionally, the need for standardized guidelines was emphasized.
Variant reinterpretation was also a topic of discussion at the 2016 Ontario Genetics Secretariat Planning Retreat Laboratory Subgroup breakout session. During that discussion, it was determined that all of the laboratories represented at the meeting reinterpret variants upon request, but will decide whether to issue an amended report on a case-by-case basis. Importantly, it was noted that at this time, some laboratories do not proactively contact referring providers if they become aware of a variant classification change. The group recognized the need for developing practice guidelines for variant reinterpretation, and emphasized the considerations and complexities of including variant reinterpretation in routine workflow of the hospital-based diagnostic laboratories that are already faced with many challenges including rising laboratory costs and staffing pressures.
Approach to reinterpretation of sequence variants
Based on our discussions with other genetic diagnostic laboratories, clinical geneticists, and genetic counselors, we have developed the following approach for variant reinterpretation (see Figure 1) in our laboratory:
-
1
Previously reported variants are not systematically reviewed.
-
2
Variant reinterpretation will be performed upon prompting. Common indications for variant reinterpretation include, but are not limited to:
-
a
a referring provider requests that the variant previously reported in their patient be reviewed, or
-
b
we receive a sample for variant-specific testing, and >12 months have elapsed since the previous variant interpretation was done; or
-
c
the variant is identified in an unrelated proband, and >12 months have elapsed since the previous variant interpretation.
-
a
-
3
If variant reinterpretation leads to a clinically significant change in classification, a laboratory genetic counselor will contact the referring provider(s) of patients who were previously found to have the same variant, or were tested for the variant as part of familial cascade testing. The updated information will be discussed, and an amended report will be issued. Note that we define a “clinically significant” change as one that may impact patient care or indication for familial cascade testing, such as:
-
a
likely benign or variant of uncertain significance upgraded to likely pathogenic or pathogenic; or
-
b
pathogenic or likely pathogenic variant downgraded to a variant of uncertain significance, likely benign or benign.
-
a
Discussion
The variant reinterpretation protocol outlined above was implemented at our institution on 1 April 2016. Since then, we have had several cases where variant reinterpretation led to a clinically significant change in classification (see Supplementary Materials). While these cases may lead to difficult genetic counseling scenarios, in our experience the providers and patients involved have been pleased to receive updated information about their genetic status, regardless of whether it was specifically requested.
Our experience suggests that developing practice guidelines regarding variant reinterpretation is necessary to ensure accurate and consistent interpretation of clinical significance of variants. We have estimated that the average time required for reinterpretation of a single variant is 30 minutes, but the actual time can vary significantly. For example, a variant that has not been reported in the literature or in patient or control population databases may take only a few minutes to review and reinterpret. However, a variant with several entries in patient databases, with differing classifications of significance and conflicting data in the literature, could take several hours to reinterpret.
While this workload is manageable in our current laboratory setting, it is likely to increase significantly in the future given the trend toward larger diagnostic panels and unbiased testing approaches (e.g., exome sequencing), and the proportional rise in variant numbers. Some commercial laboratories have implemented billing policies for variant reinterpretation, as an attempt to offset the associated costs for this work. Another option worthy of consideration is to minimize the staff time required for communication of updated variant interpretation, such as the establishment of an online database or communication portal where variant reclassification information could be automatically sent to the relevant health-care providers.
Our results demonstrate that variant reinterpretation is required to maintain the highest standards of clinical care, and highlight the importance of considering resources and staffing required for continuous variant reinterpretation when launching new next-generation sequencing–based tests in a diagnostic setting. We hope that this study will serve as a starting point and/or contribute to development of expert standards and recommendations that are required to ensure a consistent and robust approach to variant reinterpretation in a diagnostic laboratory setting.
References
Richards S, Aziz N, Bale S, Bick D, Das S . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Amendola LM, Jarvik GP, Leo MC et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet 2016;98:1067–1076.
Otten E, Plantigna M, Birnie E et al. Is there a duty to recontact in light of new genetic technologies?: a systematic review of the literature. Genet Med 2015;17:668–678.
Carrieri D, Lucassen AM, Clarke AJ et al. Recontact in clinical practice: a survey of clinical genetics services in the United Kingdom. Genet Med 2016;18:876–881.
Carrieri D, Dheensa S, Doheny S et al. Recontacting in clinical practice: an investigation of the views of healthcare professionals and clinical scientists in the United Kingdom. Eur J Hum Genet 2017;25:275–279.
Matthijs G, Souche E, Alders M et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet 2016;24:2–5.
Acknowledgements
We thank the patients, without whom this work would not be possible. We also thank all of the staff of the Regional Genetics Program at Children’s Hospital of Eastern Ontario, in particular the molecular genetics laboratory section staff, for their contributions to this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary material is linked to the online version of the paper at
Supplementary information
Rights and permissions
About this article
Cite this article
Chisholm, C., Daoud, H., Ghani, M. et al. Reinterpretation of sequence variants: one diagnostic laboratory’s experience, and the need for standard guidelines. Genet Med 20, 365–368 (2018). https://doi.org/10.1038/gim.2017.191
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.191
Keywords
This article is cited by
-
Equity implications of patient-initiated recontact and follow-up in clinical genetics
European Journal of Human Genetics (2023)
-
The Need to Standardize the Reanalysis of Genomic Sequencing Results: Findings from Interviews with Underserved Families in Genomic Research
Journal of Bioethical Inquiry (2023)
-
Assessing parental understanding of variant reclassification in pediatric neurology and developmental pediatrics clinics
Journal of Community Genetics (2021)
-
A pediatric perspective on genomics and prevention in the twenty-first century
Pediatric Research (2020)
-
Reinterpretation, reclassification, and its downstream effects: challenges for clinical laboratory geneticists
BMC Medical Genomics (2019)