Abstract
Purpose
Mutations in genes involved in Fanconi anemia (FA)/BRCA DNA repair pathway cause cancer susceptibility diseases including familial breast cancer and Fanconi anemia (FA). A single FA patient with biallelic FANCM mutations was reported in 2005 but concurrent FANCA pathogenic mutations precluded assignment of FANCM as an FA gene. Here we report three individuals with biallelic FANCM truncating mutations who developed early-onset cancer and toxicity to chemotherapy but did not present congenital malformations or any hematological phenotype suggestive of FA.
Methods
Chromosomal breakages, interstrand crosslink sensitivity, and FANCD2 monoubiquitination were assessed in primary fibroblasts. Mutation analysis was achieved through Sanger sequencing. Genetic complementation of patient-derived cells was performed by lentiviral mediated transduction of wild-type FANCM complementary DNA followed by functional studies.
Results
Patient-derived cells exhibited chromosomal fragility, hypersensitivity to interstrand crosslinks, and impaired FANCD2 monoubiquitination. We identified two homozygous mutations (c.2586_2589del4; p.Lys863Ilefs*12 and c.1506_1507insTA; p.Ile503*) in FANCM as the cause of the cellular phenotype. Patient-derived cells were genetically complemented upon wild-type FANCM complementary DNA expression.
Conclusion
Loss-of-function mutations in FANCM cause a cancer predisposition syndrome clinically distinct from bona fide FA. Care should be taken with chemotherapy and radiation treatments in these patients due to expected acute toxicity.
Similar content being viewed by others
Introduction
Fanconi anemia (FA) is a rare hereditary and recessive disease affecting about 1 in 100,000 births.1 FA patients exhibit a wide range of clinical features, each of incomplete penetrance, of which bone marrow failure (BMF) and cancer predisposition are the most life threatening.2 All FA patient-derived cells are extremely sensitive to DNA interstrand crosslinks (ICLs).2 ICLs are highly damaging to proliferating cells because they block DNA replication forks and, affecting both DNA strands, complicate error-free DNA repair.3 Therefore, ICL-inducing drugs are widely used in cancer therapy.3
A total of 21 genes associated with FA have been identified to date: FANCA, FANCB, FANCC, FANCD1 (BRCA2), FANCD2, FANCE, FANCF, FANCG (XRCC9), FANCI, FANCJ (BRIP1), FANCL (PHF9), FANCM, FANCN (PALB2), FANCO(RAD51C), FANCP (SLX4), FANCQ (ERCC4), FANCR (RAD51), FANCS (BRCA1), FANCT (UBE2T), FANCU (XRCC2), and FANCV (REV7).4 The products of eight of these genes (A, B, C, E, F, G, L, M) form a complex (FA core complex) with E3 ubiquitin ligase activity that, together with FANCT/UBE2T E2 conjugase, monoubiquitinates FANCD2 and FANCI dimer (I/D complex).2 I/D complex monoubiquitination is a key molecular event in FA pathway and defects in any of core complex–forming proteins or in UBE2T/FANCT (upstream genes) abolish this occurence.2, 5 Remaining FA genes are classified as I/D or downstream because cells deficient in these genes are able to monoubiquitinate the I/D complex despite having an FA phenotype.2
FANCM was identified in 2005 as a member of FA core complex and as the mutated gene in a FA patient.6 However, phenotype of patient-derived cells could not be complemented by wild-type (wt) FANCM complementary DNA (cDNA).6 Later it was discovered the patient also presented biallelic mutations in FANCA and a sibling with FA was actually subtyped FA-A. The two siblings did not share biallelic mutations in FANCM indicating FA phenotype in both patients was actually caused by FANCA mutations.7
Despite the fact that FANCM is not a bona fide FA gene, FANCM relevance in DNA damage response and repair is unquestionable.8 FANCM deleterious variants are associated with breast cancer susceptibility in monoallelic mutation carriers,9, 10 and FANCM has a fundamental role in recruiting Bloom complex (BLM-TOPOIIIα-RMI1-RMI2) and in remodeling stalled DNA replications forks.11 Although FANCM is not essential for RAD51 foci formation and homologous recombination repair, FA-M cells are sensitive to Camptothecin and PARP inhibitors resembling FA-D1, FA-N, FA-O, and FA-S cells.7, 12 Among FA downstream components, BRCA2/FANCD1, BRCA1/FANCS, BRIP1/FANCJ, PALB2/FANCN, RAD51C/FANCO are important genes for familiar breast and ovary cancer predisposition in monoallelic mutation carriers.2 Individuals with biallelic mutations in RAD51C/FANCO and BRCA1/FANCS are clinically classified as FA-like as they present with FA-related birth defects, chromosome fragility, or cancer but normal hematology.2
Here we describe for the first time three individuals (EGF255, EGF280, and EGF291) with biallelic truncating mutations in FANCM that developed early-onset cancer but did not present congenital malformations or any hematological phenotype suggestive of FA. Patient-derived cells presented chromosome fragility, ICL sensitivity, and cell cycle alterations typical of an FA cellular phenotype. These abnormalities were fully corrected by expression of wt FANCM cDNA thus unequivocally indicating FANCM as the disease-causing gene.
Materials and methods
Patients’ phenotype description
Patient EGF255 was a male diagnosed with B-cell precursor lymphoblastic leukemia at 14 years old. He had no personal or familial disease history and no physical signs suggestive of FA. Since he experienced major toxicity after first-line therapy for B-cell precursor lymphoblastic leukemia, i.e., intense mouth toxicity with deep and persistent pancytopenia, an underlying genetic cause was suspected. A diagnosis of FA was established using primary fibroblasts from a skin biopsy (not shown). The patient reached clinical remission but relapsed 5 years after the first diagnosis.
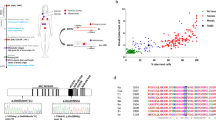
Patient EGF291, was born to a third-degree consanguineous family originating from La Réunion. He was a never-smoker but was diagnosed at age 45 with a squamous cancer of left arytenoid. He was 158 cm tall and had no history of disease. Blood counts at the time of cancer diagnosis were in normal range. He experienced a strong toxicity to chemotherapy and radiotherapy and died from hemorrhage on the progressing hypervascular larynx tumor. His younger brother, patient EGF280, who was 160 cm tall without malformations, developed a squamous cell carcinoma in the mouth at 32 years old. He had frequent exposure to tobacco and alcohol. He also presented a strong toxicity to cancer treatment. Based on family history of two cancers at young age in two brothers, a genetic syndrome was hypothesized. A chromosome breakage test was performed in peripheral blood of patient EGF280 that showed a strong excess of breaks and radials upon mitomycin C (MMC) exposure (not shown). A skin biopsy was obtained from the two brothers and primary fibroblasts showed an FA core deficient pattern by FANCD2 immunoblotting and a strong hypersensitivity to MMC (Figure 1a).
Molecular and cellular diagnostic data of the three patients. (a) Western blot demonstrated a Fanconi anemia (FA) core pattern in primary fibroblasts with lack of FANCD2-L isoform (indicated by *) after interstrand crosslinks treatment. Control FA and no-FA samples are also shown. (b) Graphs representing the mitomycin C (MMC) sensitivity data in the patients’ cells, and in FANCA, FANCG and FANCI mutated FA cells used as positive controls and non-FA cells used as negative controls. (c) and (d) Electropherograms showing the FANCM mutations in patients EGF255 and EGF280. (e) Pedigree of EGF280 and EGF291’s family. III8’s cause of death was unrecorded. Father (II3) developed a mouth cancer at unknown age. He was a smoker. III2 was found heterozygous for the c.1506_1507insTA; p.Ile503* mutation in blood and mouth brush genomic DNA. (f) Complementation of EGF255 primary fibroblasts interstrand crosslink sensitivity by transduction of wild-type FANCM complementary DNA; N = 2; error bars: STD. DEB, diepoxybutane.
Cell lines, MMC sensitivity, and immunoblots
Chromosomal breakages, MMC sensitivity, and FANCD2 monoubiquitination were assessed in primary fibroblasts from the three patients as previously described.13, 14 Mutation analysis was achieved throughout Sanger DNA sequencing of all FA core complex genes. Primary fibroblasts were immortalized by transduction of SV40LT cDNA (from pBABE-puro SV40LT) BamH1-cloned in pULTRA lentiviral vector (Addgene, Cambridge, MA, USA). pBABE-puro SV40LT was a gift from Thomas Roberts (Addgene #13970) and pUltra was a gift from Malcolm Moore (Addgene #24129). Immortalized cells were cultivated as in ref.10. FANCM cDNA expressing lentiviral vectors were described in ref. 10. All lentiviral particles were prepared and used as in ref. 10. Expression of FANCM was achieved with Tet-Express (cat #631177, Clontech, Mountain View, CA, USA) according to manufacturer’s instructions. For immunoblot studies, the following primary antibodies were used: mouse monoclonal anti-FANCM, clone CV5.1 diluted 1:100 (MABC545, MERCK Millipore, Billerica, MA, USA), mouse monoclonal anti-Vinculin diluted 1:3,000 (ab18058, Abcam, Cambridge, UK), and mouse monoclonal anti-FANCD2 diluted 1:500 (GTX70299, Genetex, Irvine, CA, USA). For FANCD2 monoubiquitination studies, cells were treated for 16 hours with hydroxyurea at the indicated doses before immunoblot analysis. Immunoblots detection was achieved with Luminata Classico (Millipore) (Vinculin and FANCD2) and LuminataForte (Millipore) (FANCM).
Functional studies
For ICL sensitivity studies, 25,000 cells were diepoxybutane treated at indicated doses and counted after three population doublings of control cells. Cells were counted with a Z2 Coulter counter (Beckman Coulter, Brea, CA, USA). Chromosome breakage analysis was performed as in ref. 10. To measure G2/M accumulation, 50,000 cells were diepoxybutane treated at the indicated doses and processed 72 hours after by flow cytometry. Cell cycle data analysis was performed with Flowjo software (Flowjo, LLC, Ashland, OR, USA).
Results
FANCD2 immunoblotting evidenced impaired FANCD2 monoubiquitination for the three patients analyzed (Figure 1a) while a clear hypersensitivity to MMC was demonstrated using a flow cytometry–based assay14 (Figure 1b). Subsequent Sanger sequencing of all known FA core complex FANC genes from patient EGF255 identified a FANCM homozygous mutation: c.2586_2589del4; p.Lys863Ilefs*12 (Figure 1c) while no mutations were detected in any other FA core genes. A different FANCM homozygous mutation, c.1506_1507insTA; p.Ile503*, was detected in brothers EGF280 and EGF291 (Figure 1d). The mutation was also found in EGF280 peripheral blood mononuclear cells. A third brother, born in 1963, was doing well in 2013. He was found heterozygous for the c.1506_1507insTA; p.Ile503* mutation in blood and mouth brush genomic DNA. Two more sisters were doing well, but an additional brother died at birth for unknown reasons (Figure 1e). No genetic information could be obtained for these siblings.
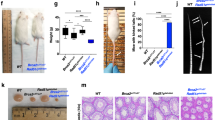
To unequivocally verify FANCM as the disease-causing gene in the two families, we performed genetic complementation by wt FANCM cDNA transduction. EGF255 primary fibroblasts sensitivity to ICLs could be complemented by expressing wt FANCM cDNA, indicating FANCM deficiency as responsible for EGF255 cellular phenotype (Figure 1f). Given the poor growth rate of the primary fibroblasts (almost 3 weeks for one population doubling), additional complementation analysis experiments were done in SV40-transformed fibroblasts from EGF255 and EGF280 patients. As shown in Figure 2a and Figure 2i–l, no FANCM band is detected in immunoblots of EGF255 and EGF280 transformed fibroblasts and FANCM cDNA transduction restored FANCM expression. As an internal control we used previously described FANCM mutation c.5791C>T (Δ22FANCM), which produces skipping of exon 22 leading to a prematurely truncated FANCM protein (p.Gly1906Alafs12*) that lacks ERCC4-like and FAAP24-interacting domains.10, 11 Only wt FANCM rescued ICLs hypersensitivity, chromosome fragility, cell cycle arrest, and FANCD2 monoubiquitination of both EGF255 and EGF 280 transformed cell lines (Figure 2c–l). Of interest, DNA repair was not completely abolished in cells expressing Δ22 FANCM as expression of this mutant form resulted in an intermediate cellular phenotype (Figure 2c–h), suggesting that the p.Gly1906Alafs12* mutation is actually hypomorphic. This result could probably explain milder clinical features of patients with this mutation in both alleles (see the related paper by Catucci et al.15).
Genetic complementation analysis of EGF255 and EGF280 immortalized fibroblasts. (a) and (b) Immunoblots showing expression of FANCM in the EGF255 and EGF280 immortalized fibroblasts used in the study. FANCM p.Gly1906Alafs12* form (Δ22FANCM) lacking exons 22 and 23 appears in the immunoblot as a shorter band. (c) Complementation of EGF255 immortal cell line ICLs sensitivity by transduction of wild-type (wt) FANCM complementary DNA. Transduction of the truncated form Δ22FANCM does not normalize interstrand crosslink (ICL) sensitivity while wtFANCM fully restores resistance to ICLs; N = 2; error bars: STD. (d) Chromosome fragility analysis of the cell lines used in (c). (e) G2/M Block analysis after treatment with ICLs of the same cell lines used in (c) and (d); N = 2; error bars: STD. (f) Complementation of EGF280 immortal cell line ICLs’ sensitivity by transduction of wt FANCM cDNA. Transduction of the truncated form Δ22FANCM does not normalize ICLs’ sensitivity while wtFANCM fully restores resistance to ICLs; N = 2; error bars: STD. (g) Chromosome fragility analysis of the cell lines used in (f). (h) G2/M Block analysis after treatment with ICLs of the same cell lines used in (f) and (g); N = 2; error bars: STD. (i) Immunoblot showing that only the transduction of wt FANCM fully rescues hydroxyurea (HU)-induced FANCD2 monoubiquitination in EGF255 immortalized fibroblasts. (j) Immunoblot showing that only the transduction of wt FANCM fully rescues HU-induced FANCD2 monoubiquitination in EGF280 immortalized fibroblasts. DEB, diepoxybutane.
Discussion
Our data suggest that biallelic truncating FANCM mutations cause an FA-like early-onset cancer syndrome rather than BMF. Patients developed either B-cell precursor lymphoblastic leukemia (EGF255) or head-and-neck carcinoma (EGF280 and EGF291) and did not present BMF or birth defects reminiscent of FA. None of the eight cancer patients with biallelic FANCM mutations reported here and in the related paper by Catucci et al.15 presented BMF or any other FA-related malformation even if the age of all patients is far beyond the mean age of hematological onset reported for bona fide FA patients. Our patients’ cells showed impairment of FANCD2 monoubiquitination (Figure 1a and Figure 2i–l) while FANCM loss in mice and chicken cells leads to reduced FANCD2 monoubiquitination.16, 17 This can be explained by our cells’ low proliferation rates, which can affect FANCD2 monoubiquitination since it occurs only in S-phase.18 Our patients’ primary fibroblasts needed almost three weeks for one population doubling and roughly a week was necessary for their immortalized counterparts. Indeed, in Figure 2l, a faint FANCD2 monoubiquitinated band is present after hydroxyurea induction in the immortalized FANCM-deficient cells.
Biallelic FANCD1/BRCA2 and FANCN/PALB2 mutations produce severe forms of FA characterized by acute BMF, congenital birth defects, chromosomal instability and occurrence of leukemia and multiple tumors usually in the first decade of life. Cancer spectrum in FA-D1 and FA-N patients is wider than in FA patients harboring mutations in other FA genes, and head-and-neck carcinoma has not been reported in a cohort of 31 FA-D1 patients.19 Biallelic mutations in FANCO/RAD51C and FANCS/BRCA1 produce an FA-like syndrome with FA-related birth defects and chromosomal instability but no BMF. FA-S patients also develop breast and ovary cancer at adult age20 while no FA-O patients with cancer have been reported so far.2 Our male patients developed typical FA malignancies like acute lymphocytic leukemia and head-and-neck carcinoma at young age. Acute lymphocytic leukemia is not a common FA-related cancer, but is far more frequent in FA patients (0.66%)21 than in the general population (0.0017%) (https://seer.cancer.gov/statfacts/html/alyl.html). Hitherto, individuals with FANCM variants were reported susceptible only to breast cancers. This suggests the working hypothesis that FANCM mutations produce different phenotypic manifestations (related paper by Catucci et al.15) when affecting different FANCM domains. Patient EGF255 harbors an FANCM truncation at the amino acid 503 and in patients EGF280 and EGF291 FANCM is truncated at the amino acid 863 but we could not detect any extra bands in our immunoblots (not shown). Since the mouse monoclonal antibody used in our study was raised against whole FANCM, we cannot rule out if the truncated FANCM proteins are physiologically stable since the epitope could be in the missing C-terminus part. Even if the truncated FANCM proteins would be expressed, they would lack all domains responsible for the interactions with Bloom syndrome complex and with FAAP24 in the FA core complex.11 In a recent study that identified loss-of-function variants in the Finnish population, at least seven probands with biallelic truncating mutations in FANCM were found without any clinical phenotype. Two truncating mutations were identified: c.5101C>T (p.Gln1701*) and c.5791C>T (p.Arg1931*) altering only the FANCM FAAP24 interaction domain.11 It is thus possible that truncating mutations that affect multiple aspects of FANCM’s role in DNA damage response and repair contribute to the distinctive phenotype observed. This is further supported by the fact that Fancm−/− mouse model phenotype recapitulates the human one: mice completely lacking Fancm are cancer prone but do not develop any BMF.16 In conclusion, severe biallelic truncating mutations in FANCM do not lead to FA but cause an FA-like cancer predisposition syndrome. Care should be taken when treating these patients with chemotherapy and radiation due to expected acute toxicity in bone marrow and other tissues.
References
Rosenberg PS, Tamary H, Alter BP . How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi anemia in the United States and Israel. Am J Med Genet A 2011;155A:1877–1883.
Bogliolo M, Surralles J . Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev 2015;33:32–40.
Deans AJ, West SC . DNA interstrand crosslink repair and cancer. Nat Rev Cancer 2011;11:467–480.
Bluteau D, Masliah-Planchon J, Clairmont C et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest 2016;126:3580–3584.
Hira A, Yoshida K, Sato K et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet 2015;96:1001–1007.
Meetei AR, Medhurst AL, Ling C et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 2005;37:958–963.
Singh TR, Bakker ST, Agarwal S et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 2009;114:174–180.
Xue X, Sung P, Zhao X . Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev 2015;29:1777–1788.
Kiiski JI, Pelttari LM, Khan S et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc Natl Acad Sci USA 2014;111:15172–15177.
Peterlongo P, Catucci I, Colombo M et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum Mol Genet 2015;24:5345–5355.
Deans AJ, West SC . FANCM connects the genome instability disorders Bloom’s syndrome and Fanconi anemia. Mol Cell 2009;36:943–953.
Stoepker C, Faramarz A, Rooimans MA et al. DNA helicases FANCM and DDX11 are determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2015;26:54–64.
Soulier J, Leblanc T, Larghero J et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood 2005;105:1329–1336.
Pinto FO, Leblanc T, Chamousset D et al. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica 2009;94:487–495.
Catucci I, Osorio A, Arver B et al. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med 2017;19:xxx–xxx.
Bakker ST, van de Vrugt HJ, Rooimans MA et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet 2009;18:3484–3495.
Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ . The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res 2009;37:4360–4370.
Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD . S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 2002;100:2414–2420.
Meyer S, Tischkowitz M, Chandler K, Gillespie A, Birch JM, Evans DG . Fanconi anaemia, BRCA2 mutations and childhood cancer: a developmental perspective from clinical and epidemiological observations with implications for genetic counselling. J Med Genet 2014;51:71–75.
Sawyer SL, Tian L, Kahkonen M et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 2015;5:135–142.
Kutler DI, Singh B, Satagopan J et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 2003;101:1249–1256.
Acknowledgements
J.S.’s laboratory is supported by the ICREA-Academia program, Spanish Ministry of Health (projects FANCOSTEM and FANCOLEN), Spanish Ministry of Economy and Competiveness (projects CB06/07/0023, SAF2012-31881 and SAF2015-64152-R), European Commission (EUROFANCOLEN project HEALTH-F5-2012-305421, FANCODRUG project H2020-703521 and P-SPHERE COFUND project), Fanconi Anemia Research Fund, and “Fondo Europeo de Desarrollo Regional, una manera de hacer Europa” (FEDER). CIBERER is an initiative of Instituto de Salud Carlos III, Spain. We thank Cristian Valiente for technical help with FANCM western blots.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
The first two authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Bogliolo, M., Bluteau, D., Lespinasse, J. et al. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet Med 20, 458–463 (2018). https://doi.org/10.1038/gim.2017.124
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.124
Keywords
This article is cited by
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
FANCA deficiency promotes leukaemic progression by allowing the emergence of cells carrying oncogenic driver mutations
Oncogene (2023)
-
Fanconi Anemia Gene Variants in Patients with Gonadal Dysfunction
Reproductive Sciences (2022)
-
DONSON and FANCM associate with different replisomes distinguished by replication timing and chromatin domain
Nature Communications (2020)
-
The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT)
Nature Communications (2019)