Abstract
Purpose
To elucidate the novel molecular cause in two unrelated consanguineous families with autosomal recessive intellectual disability.
Methods
A combination of homozygosity mapping and exome sequencing was used to locate the plausible genetic defect in family F162, while only exome sequencing was followed in the family PKMR65. The protein 3D structure was visualized with the University of California–San Francisco Chimera software.
Results
All five patients from both families presented with severe intellectual disability, aggressive behavior, and speech and motor delay. Four of the five patients had microcephaly. We identified homozygous missense variants in LINGO1, p.(Arg290His) in family F162 and p.(Tyr288Cys) in family PKMR65. Both variants were predicted to be pathogenic, and segregated with the phenotype in the respective families. Molecular modeling of LINGO1 suggests that both variants interfere with the glycosylation of the protein.
Conclusion
LINGO1 is a transmembrane receptor, predominantly found in the central nervous system. Published loss-of-function studies in mouse and zebrafish have established a crucial role of LINGO1 in normal neuronal development and central nervous system myelination by negatively regulating oligodendrocyte differentiation and neuronal survival. Taken together, our results indicate that biallelic LINGO1 missense variants cause autosomal recessive intellectual disability in humans.
Similar content being viewed by others
Introduction
Intellectual disability (ID) is a common neurodevelopmental disorder, affecting 2% of the general population, which poses a considerable burden on the quality of life of the families of affected individuals and has a significant socioeconomic impact on society and the health-care system.1 It is an extremely heterogeneous group of disorders, as to date, over 700 genes have been implicated in syndromic and nonsyndromic ID.2 Despite the considerable progress in disease gene identification, especially after the introduction of next-generation sequencing, at least 50% of the estimated genetic causes of ID remain unknown.3 All Mendelian inheritance patterns have been reported with ID, and approximately half of them follow an autosomal recessive inheritance pattern (autosomal recessive intellectual disability).3
The study of consanguineous families has facilitated the discovery of pathogenic genetic variants in many autosomal recessive disorders.4,5,6 The frequency of autosomal recessive disorders is higher in populations where consanguineous marriages are frequent.7 Consanguinity results in higher risk for birth defects varying from 2.7% to 15.8%.8 The combination of homozygosity mapping and exome sequencing is a powerful and cost-effective tool for molecular diagnosis and discovery of novel genes in families with suspected autosomal recessive disorders.9,10,11,12,13
Using this approach, we studied two unrelated Pakistani consanguineous ID families, and identified two different homozygous missense variants in LINGO1 (leucine-rich repeat and immunoglobulin domain containing 1) that are likely to cause the intellectual disability.
Materials and methods
Patients
Family F162 was recruited and collected from the Institute of Basic Medical Sciences, Khyber Medical University, Peshawar, Pakistan, and family PKMR65 was enrolled at Allama Iqbal Medical Research Center, Lahore, Pakistan. The study was approved by the Bioethics Committee of the University Hospitals of Geneva (protocol CER 11–036); the Institutional Review Board of the Centre of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan; and the Medical Ethical Committee Arnhem-Nijmegen, The Netherlands. Written informed consent was provided by all parents or legal representatives for the performed analyses.
Exome sequencing
In case of family F162, exome capture was performed with the SureSelect Human All Exon kit v5 (Agilent Technologies, Santa Clara, CA). The exome-captured library was sequenced on an Illumina HiSeq2000 with 125-bp paired-end reads yielding an average of 220 × coverage per targeted base. Exome-sequencing data was analyzed using an in-house customized pipeline. The pipeline is based on published algorithms including the Burrows–Wheeler aligner tool (BWA),14 SAMtools,14 Pindel,15 and ANNOVAR.16 The pipeline uses these algorithms to map the reads (BWA), detect variants (SAMtools) and indels (Pindel), and the annotation (ANNOVAR) in a sequential manner. To calculate coverage and on-target reads for the entire coding sequence, human RefSeq17 coding genes were used as the reference.
To identify the causative variant in the family PKMR65, exome sequencing was performed using similar reagents and platform as for family F162, but with 100-bp paired-end reads. Approximately 95% of the reads were mapped uniquely to the reference sequence with a mean coverage of 125 ×. The reads were aligned to the reference human genome (GRCh37/hg19), using the BWA.14 Polymerase chain reaction duplicates were identified using Picard (http://broadinstitute.github.io/picard).
Homozygosity mapping
Homozygosity mapping was performed in family F162 by 720-K single-nucleotide polymorphism (SNP) array (HumanOmniExpress Bead Chip by Illumina, San Diego, CA) with average SNP density of one SNP per 4 kb and a window of 50 consecutive homozygous SNPs, with maximum of one mismatch allowed in each homozygous region. The runs of homozygosity (ROH) regions were demarcated by the first heterozygous SNPs adjoining each homozygous region. Potential target genomic regions were identified as the ROHs shared by the affected individuals, but not by their parents or their unaffected siblings.
Bioinformatics analysis
For family 162, the software CATCH18 was used to analyze consanguineous families by combining family pedigree information, ROH, and exome-sequencing data. This software automatically marks homozygous variants as “putative” if they are present in ROHs of patients but not in unaffected individuals of a nuclear family. Subsequently, the variants were filtered manually using the criteria described in previous studies.11 As an initial filter in the target region of each family, we included all homozygous exonic and splicing variants (±6 bp of the intron–exon boundary) and excluded all synonymous variants, which are not in the splicing regions. We selected variants with a minor allele frequency <0.02 in the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org) and our local database, and the variants found within duplications of the genome were also filtered out. After the initial filtering, all remaining variants were evaluated and ranked on the basis of conservation (by GERP++);19 predicted pathogenicity scores such as by SIFT,20 PolyPhen2,21 and Mutation Taster;22 and the presence of variants in the professional version of the Human Gene Mutation Database.23 Illumina GenomeStudio software (http://www.illumina.com/software/genomestudio_software.ilmn) and the in-house built program CoverageMaster (F. Santoni, unpublished data) were used to perform the copy-number variation (CNV) analysis using SNP-array and exome-sequencing coverage data, respectively. The final list of variants was further verified by Sanger sequencing in all family members whose DNA samples were available to determine whether they segregated with the disease phenotype.
The exome sequencing data of family PKMR65 were analyzed by calculating the coverage and on-target reads for the entire coding sequence. CNV analysis on the exome data was performed following the method described previously.24 Next, the selection of variants was performed as described previously.25 In short, seven major steps were taken to select all high-quality potentially pathogenic variants: (i) inclusion of variants present in at least four reads and present in ≥80% of all reads; (ii) exclusion of those homozygous variants that are present in unaffected controls sequenced at the same time; (iii) exclusion of variants within intergenic, intronic (apart from the splice site variants), and untranslated regions; (iv) exclusion of variants present in dbSNP142, 1000 Genome, the National Heart, Lung, and Blood Institute Exome Variant Server database, or ExAC database with a frequency ≥1% (v) inclusion of loss-of-function variants (i.e., nonsense, frameshift, and splice site mutations) with a phyloP score ≥0; (v) inclusion of missense variants and in-frame deletions and duplication with a combined annotation dependent depletion score of ≥20; (vi) selected variants in genes that are expressed in the brain based on their expressed sequence tag profile in the Unigene database (transcripts per million ≥5); and (vii) inclusion of variants that segregate with the disease in the respective pedigree as determined using Sanger sequencing.
Molecular modeling analysis
The crystal structure of the LINGO1 protein tetramer stored under the 2ID5 code in the Protein Data Bank (http://www.rcsb.org/) was used for molecular modeling studies.25 The influence of the residues on the structural stability was studied by computational alanine scanning using the Foldx4 software.26 The structural stability of the wild-type protein was computed and compared with that of the alanine mutants. The stability is estimated as a difference between the free energy of the unfolded and folded states of the protein. The change in the stability upon mutation to alanine indicates the importance of each residue side chain for the structure of the LINGO1 protein.27 The protein was visualized with the University of California–San Francisco Chimera software.27
Results
Clinical evaluation
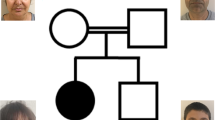
Family F162 (Figure 1a) has two affected individuals (IV:1 and IV:8) and six unaffected siblings (IV:3, IV:4, IV:5, IV:6, IV:9, and IV:10), the parents of whom are first cousins. One of the siblings (IV:2) died two days after birth, the cause of which is not known, and one stillbirth was also reported in this family. Both affected individuals of this family were clinically evaluated. At the time of clinical evaluation, age of female (IV:1) and male (IV:8) affected individuals was 10.5 and 21 years, respectively. They presented with severe ID, microcephaly, developmental delay, aggressive behavior, and slurred speech. Affected individuals (IV:1 and IV:8) started walking at the age of 2 and 10 years, respectively. In addition to these phenotypes, one of the patients (IV:8) also had uncontrolled epilepsy. Brain magnetic resonance imaging of both the affected individuals (IV:1 and IV:8) did not reveal any signs or symptoms of obvious myelination defects.
Family PKMR65 with probable autosomal recessive intellectual disability consisted of three affected female individuals (VI:2, VI:3, and VI:6) and three unaffected siblings (VI:1, VI:4, and VI:5) born to consanguineous parents. At the time of evaluation, affected individuals VI:2, VI:3, and VI:6 were aged 25, 20, and 7 years, respectively. Two affected individuals (VI:2 and VI:3) had severe and one affected individual (VI:6) had moderate intellectual disability. All childhood developmental milestones (gross motor, fine motor, speech, and social) were delayed. Affected individuals could not speak, and had poor social interaction and aggressive labile mood. Individuals VI:2 and VI:3 with severe ID also could not eat or drink independently. Individual VI:2 had a history of left hemiparesis at 9 years of age that had recovered over time and medical record was not available. There was no history of epilepsy.
Physical examination was remarkable for spastic hypertonia and exaggerated deep tendon reflexes without motor deficit in two individuals (VI:2 and VI:3). Two individuals (VI:3 and VI:6) were microcephalic. Except for a left dysplastic ear in individual VI:6, no other dysmorphic features were observed. Fundoscopic examination of all affected individuals was unremarkable. No brain magnetic resonance images were available for the affected individuals of family PKMR65. A considerable phenotypic concordance was found among all five affected individuals of both families, as summarized in Table 1.
Genetic analysis
The SNP-array analysis was performed to genotype six individuals (III:4, III:5, IV:1, IV:3, IV:6, and IV:8) of family F162, including both affected individuals, their unaffected siblings, and both parents. ROHs were calculated using PLINK25. Family structure was verified by calculating identity by descent through PLINK25 to estimate the relatedness among individuals of the family. Subsequently, exome sequencing was performed in one of the affected individuals (IV:8) of family F162. A total of 21,739 high-quality exonic and 1,302 canonical splice site (within ±6 nucleotides from the intron–exon boundary) variants were found, covering 97.2% of the target regions with at least 10 × resolution. The CATCH18 analysis resulted in two candidate variants (NM_032808.6:c.869G>A:p.(Arg290His) in LINGO1 (MIM 609791) and NM_004998.3:c.1406C>T:p.(Thr469Met) in MYO1E (MIM 601479)), which were present in the same ROH, segregating with the disease phenotype. The segregation of stated genotypes with the ID phenotype was confirmed by Sanger sequencing in all available family members in family F162 (Figure 1a). As mutations in MYO1E are known to cause autosomal recessive nephrotic syndrome (OMIM 601479)28 and the affected individuals did not show any symptoms of nephrotic syndrome, the variant found in MYO1E appears not pathogenic. To confirm this, we tested the excretion of albumin in urine samples, as nephrotic syndrome causes the excretion of high level of proteins in the urine,29 showing no secretion of albumin in the urine of either patient. Moreover, clinical examination of the affected individuals did not reveal edema or swelling of hand or face, confirming that the MYO1E variant is benign.
By sharing the data with collaborating investigators, another consanguineous family PKMR65 segregating a likely pathogenic variant (NM_032808.6:c.863 A>G: p.(Tyr288Cys)) in the LINGO1 gene was identified. Exome sequencing of VI:2 and VI:3 of family PKMR65 identified candidate missense variants in DNAJC2, LINGO1, and VAPA.25 Sanger sequencing showed that only the c.863 A>G (p.(Tyr288Cys)) in LINGO1 segregated with the ID phenotype in the family (Figure 1b), pointing to LINGO1 as the prime ID candidate gene in PKMR65. CNV analysis using SNP-array data of family F162 or exome-sequencing data of both families (F162 and PKMR65) did not reveal any likely pathogenic CNV.
Neither of the LINGO1 variants (NM_032808.6:c.869 G>A:p.(Arg290His)) and (NM_032808.6:c.863A>G:p.(Tyr288Cys)) found in families F162 and PKMR65, respectively, were present in the ethnically matched control cohort (n = 201) or in the South Asian population in the GnomAD database (Supplementary Table 1 online).
Molecular modeling analysis
In humans, at least 12 different isoforms of LINGO1 are expressed, which vary from each other at their 5′ untranslated regions. The coding exons of LINGO1 are identical for each isoform and encode a polypeptide of 620 amino acids that belongs to a large family of leucine-rich repeat immunoglobulin domains containing proteins. The extracellular amino terminal region known as the ectodomain has a tandem array of multiple leucine-rich repeats and a single immunoglobulin-like domain followed by a transmembrane domain and short cytoplasmic tail.25 Both mutant amino acid residues, at Arg290 and Tyr288, are located in the leucine-rich repeat region of LINGO1 and are conserved down to zebrafish (Figure 2). The leucine-rich repeat region of LINGO1 is thought to be essential for protein–protein interactions. Therefore, it is possible that the substitutions will affect binding of LINGO1 to other proteins. Visual inspection of the crystal structural model of the protein revealed that both mutated amino acid residues are close to a glycosylation site at position Asn264, suggesting that they may interfere with proper glycosylation (Figure 3). The estimated stability change upon alanine mutation were nonsignificant for both residues: 0.1 kcal/mol in the case of Arg290 and 0.3 kcal/mol loss in the case of Tyr288, suggesting negligible influence of both residues on the protein stability. This is not surprising since both residues are located on the protein surface, pointing toward the solution. Nevertheless, it cannot be excluded that the p.(Tyr288Cys) mutation is influencing the folding of LINGO1 as it introduces additional free thiol groups that can react during folding and create wrong disulfide bridges.
(a) A visual inspection of the structure suggests that both variants might interfere with glycosylation process or the recognition of the glycosylation site. Yellow spheres represent p.Arg290 and p.Tyr288, whereas glycan chain is in red and other glycan chains are also shown in sphere representation. (b) Tetrameric structure of lingo-1. Glycan on residue 264 not shown. The protein was visualized with the University of California–San Francisco Chimera software.
Discussion
We have identified two homozygous missense variants in LINGO1, p.(Arg290His) and p.(Tyr288Cys), that both fully segregated with the neurodevelopmental phenotype in family F162 and family PKMR65. All patients from both families presented with a similar phenotype consisting of severe intellectual disability, aggressive behavior, speech delay, and motor delay. Four of the five patients had microcephaly. One patient from family F162 presented with epilepsy. Brain magnetic resonance imaging on both affected individuals (IV:1 and IV:8) of family F162 did not detect structural abnormalities or myelination defects. The LINGO1 locus (15q24-26) has previously been associated with autism, schizophrenia, anxiety, and depression, and alterations in this region have been implicated as a susceptible factor for psychiatric disorders,30 supporting a role for this gene in neurodevelopmental disorders. The two mutated amino acid residues in LINGO1, at Arg290, and Tyr288 are conserved down to zebrafish (Figure 2).
It is interesting to note that LINGO1 may tolerate neither loss-of-function nor missense variants. Data from ExAC31 show that only half the expected number of missense variants have been detected (Z value of 4.00) and that the probability of loss-of-function intolerance score has been estimated to be 0.95 showing a considerable intolerance to loss-of-function variants. These metrics are usually found in genes that are involved in severe autosomal dominant disorders with reduced fecundity such as ID. However, autosomal recessive modes of inheritance may also be possible under the assumption that each of the identified variants is a hypomorph and does not fully abolish protein function. This suggests that de novo LINGO1 loss-of-function variants may be observed in the future in patients with dominant ID. Alternatively, these mutations might be incompatible with life.
LINGO1 is a transmembrane protein that is predominantly expressed in the central nervous system (CNS), especially in the oligodendrocytes and neuronal cells. It has been demonstrated that LINGO1 is part of the LINGO1–RTN4R/NGFR receptor complex that negatively regulates myelination, oligodendrocyte differentiation, axon regeneration, and neuronal survival.32,33 Increased LINGO1 expression has been found in various animal models with CNS injury and in the CNS diseases in humans.34 In transgenic mice, overexpression of LINGO1 causes reduction in myelination, and less-differentiated oligodendrocytes were observed.35 Overexpression of LINGO1 is also observed in several neuronal disorders including multiple sclerosis, Parkinson disease, and essential tremor.36 Various association studies have also shown the role of LINGO1 variants with essential tremor and Parkinson’s disease.37 Moreover, inhibitory anti-LINGO1 antibodies promoted illegitimate oligodendrocyte differentiation and myelination of neurons by inhibiting the function of LINGO1. A similar outcome was observed in the Lingo1 knockout mice and zebrafish lingo1b knockdown models, which revealed early improvement in neuronal myelination as compared to wild type.38,39 Taken together, these studies establish an essential role for LINGO1 in myelination, neuronal survival, and CNS repair in general.
In conclusion, we show that homozygous LINGO1 missense variants cause severe autosomal recessive intellectual disability, aggressive behavior, speech delay, motor delay, and microcephaly. Further functional studies are warranted to dissect the exact pathophysiological mechanism of this new autosomal recessive ID syndrome.
References
Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil 2011;32:419–436.
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2016;17:9–18.
Willemsen MH, Kleefstra T. Making headway with genetic diagnostics of intellectual disabilities. Clin Genet 2014;85:101–110.
Waryah AM, Narsani AK, Sheikh SA, Shaikh H, Shahani MY. The novel heterozygous Thr377Arg MYOC mutation causes severe juvenile open angle glaucoma in a large Pakistani family. Gene 2013;528:356–359.
Zelinger L, Banin E, Obolensky A et al. A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am J Hum Genet 2011;88:207–215.
Estrada-Cuzcano A, Neveling K, Kohl S et al. Mutations in C8orf37, encoding a ciliary protein, are associated with autosomal-recessive retinal dystrophies with early macular involvement. Am J Hum Genet 2012;90:102–109.
Woods CG, Cox J, Springell K et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet 2006;78:889–896.
Zlotogora J. What is the birth defect risk associated with consanguineous marriages? Am J Med Genet 2002;109:70–71.
Iqbal Z, Willemsen MH, Papon MA et al. Homozygous SLC6A17 mutations cause autosomal-recessive intellectual disability with progressive tremor, speech impairment, and behavioral problems. Am J Hum Genet 2015;96:386–396.
Makrythanasis P, Kato M, Zaki MS et al. Pathogenic variants in PIGG cause intellectual disability with seizures and hypotonia. Am J Hum Genet 2016;98:615–626.
Makrythanasis P, Nelis M, Santoni FA et al. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum Mutat 2014;35:1203–1210.
Riazuddin S, Hussain M, Razzaq A et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry; e-pub ahead of print 26 July2016.
Najmabadi H, Hu H, Garshasbi M et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011;478:57–63.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865–2871.
Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007;35:D61–D65.
Santoni FA, Makrythanasis P, Antonarakis SE. CATCHing putative causative variants in consanguineous families. BMC Bioinformatics 2015;16:310.
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 2010;6:e1001025.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–1081.
Adzhubei IA, Schmidt S, Peshkin L et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–576.
Stenson PD, Mort M, Ball EV, Shaw K, Phillips AD, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2013;133:1–9.
de Ligt J, Boone PM, Pfundt R et al. Platform comparison of detecting copy number variants with microarrays and whole-exome sequencing. Genom Data 2014;2:144–146.
Mosyak L, Wood A, Dwyer B et al. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem 2006;281:36378–36390.
Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 2002;320:369–387.
Pettersen EF, Goddard TD, Huang CC et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–1612.
Mele C, Iatropoulos P, Donadelli R et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med 2011;365:295–306.
Hull RP, Goldsmith DJ. Nephrotic syndrome in adults. BMJ 2008;336:1185–1189.
McInnes LA, Nakamine A, Pilorge M et al. A large-scale survey of the novel 15q24 microdeletion syndrome in autism spectrum disorders identifies an atypical deletion that narrows the critical region. Mol Autism 2010;1:5.
Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291.
Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol 2008;40:1971–1978.
Mi S, Lee X, Shao Z et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci 2004;7:221–228.
Mi S, Hu B, Hahm K et al. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med 2007;13:1228–1233.
Lee X, Yang Z, Shao Z et al. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci 2007;27:220–225.
Inoue H, Lin L, Lee X et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson’s disease models. Proc Natl Acad Sci USA 2007;104:14430–14435.
Deng H, Gu S, Jankovic J. LINGO1 variants in essential tremor and Parkinson’s disease. Acta Neurol Scand 2012;125:1–7.
Yin W, Hu B. Knockdown of Lingo1b protein promotes myelination and oligodendrocyte differentiation in zebrafish. Exp Neurol 2014;251:72–83.
Mi S, Pepinsky RB, Cadavid D. Blocking LINGO-1 as a therapy to promote CNS repair: from concept to the clinic. CNS Drugs 2013;27:493–503.
Acknowledgments
We thank the Swiss Government Excellence Scholarships program, which provides M.A. the opportunity to work at the University of Geneva Medical School in Switzerland. We also thank the University Hospitals of Geneva for supporting the infrastructure for exome sequencing. The sequencing service of patient F162 IV-8’s exome was provided by the Norwegian Sequencing Center, a national technology platform supported by the Functional Genomics and Infrastructure programs of the Research Council of Norway and the Southeastern Regional Health Authorities. This project was partially supported by the Synapsy NCCR grant from the Swiss National Foundation, and ERC grant 219968. Z.I. was supported by South-Eastern Norway Regional Health Authority. The project also received funding from EU FP7 Large-Scale Integrating Project Genetic and Epigenetic Networks in Cognitive Dysfunction (241995 to H.v.B. and S.R.), and the Higher Education Commission in Pakistan. D.L.P. is recipient of a CAPES Fellowship (99999.013311/2013-01). We thank H. Venselaar, Center for Molecular and Biomolecular Informatics, Radboud University Medical Center, Nijmegen, The Netherlands, for her input in the molecular modeling of LINGO1 protein. We are also grateful to all members of the families enrolled in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ansar, M., Riazuddin, S., Sarwar, M. et al. Biallelic variants in LINGO1 are associated with autosomal recessive intellectual disability, microcephaly, speech and motor delay. Genet Med 20, 778–784 (2018). https://doi.org/10.1038/gim.2017.113
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.113
This article is cited by
-
Depression pathophysiology, risk prediction of recurrence and comorbid psychiatric disorders using genome-wide analyses
Nature Medicine (2023)
-
FOXI3 pathogenic variants cause one form of craniofacial microsomia
Nature Communications (2023)
-
Biallelic truncation variants in ATP9A are associated with a novel autosomal recessive neurodevelopmental disorder
npj Genomic Medicine (2021)