Abstract
Purpose:
Chromosomal microarray analysis (CMA) is currently considered first-tier testing in pediatric care and prenatal diagnosis owing to its high diagnostic sensitivity for chromosomal imbalances. The aim of this study was to determine the efficacy and diagnostic power of CMA in both fresh and formalin-fixed paraffin-embedded (FFPE) samples of products of conception (POCs).
Methods:
Over a 44-month period, 8,118 consecutive samples were received by our laboratory for CMA analysis. This included both fresh (76.4%) and FFPE samples (22.4%), most of which were ascertained for recurrent pregnancy loss and/or spontaneous abortion (83%). The majority of samples were evaluated by a whole-genome single-nucleotide polymorphism (SNP)-based array (81.6%); the remaining samples were evaluated by array-comparative genomic hybridization (CGH).
Results:
A successful result was obtained in 7,396 of 8,118 (91.1%), with 92.4% of fresh tissue samples and 86.4% of FFPE samples successfully analyzed. Clinically significant abnormalities were identified in 53.7% of specimens (3,975 of 7,396), 94% of which were considered causative.
Conclusion:
Analysis of POC specimens by karyotyping fails in 20–40% of cases. SNP-based CMA is a robust platform, with successful results obtained in >90% of cases. SNP-based CMA can identify aneuploidy, polyploidy, whole-genome homozygosity, segmental genomic imbalances, and maternal cell contamination, thus maximizing sensitivity and decreasing false-negative results. Understanding the etiology of fetal loss enables clarification of recurrence risk and assists in determining appropriate management for future family planning.
Genet Med 19 1, 83–89.
Similar content being viewed by others
Introduction
It is estimated that 15–20% of clinically recognized pregnancies end in miscarriage, and approximately 1% of couples experience recurrent (at least two) pregnancy loss.1,2,3 The single most common cause of early (first-trimester) pregnancy loss is the presence of a major fetal chromosomal abnormality, which is responsible for more than 50% of losses in the first trimester. Whole-chromosome aneuploidies are the most common etiology.4,5,6,7,8,9,10,11 Fetal chromosomal abnormalities account for approximately 8–10% of intrauterine fetal demises occurring after 20 weeks of gestation and/or stillbirths occurring in the second or third trimester.9,12 The etiologic analysis of pregnancy loss can provide important information for medical management, reproductive counseling, and supportive patient care.4,13,14
Although fetal karyotyping has been used for many years to evaluate samples of products of conception (POCs), there are two primary limitations to this method. First, cytogenetic analysis requires live cells, which requires culture. This is challenging for POC samples because culture failures preclude obtaining a result in approximately 20–40% of cases and lead to a prolonged turnaround time when successful.9,15,16,17,18,19 Additionally, the quality and viability of POC samples are critical for successful cell culture and karyotype analysis.4,8 Second, even when tissue is dissected carefully, there is always the risk of an erroneous result due to maternal cell contamination (MCC). Specifically, selective overgrowth of maternal cells during culture can result in a normal female karyotype even in the presence of an underlying fetal chromosome abnormality. False-negative results due to MCC have been reported to occur in approximately 29–58% of POC cases analyzed by techniques requiring the use of cell culture.16,17,18,19
High-resolution chromosomal microarray analysis (CMA) using single-nucleotide polymorphism (SNP)-based arrays has replaced karyotyping in most pediatric and prenatal diagnostic applications owing to its ability to simultaneously detect aneuploidies, submicroscopic chromosomal imbalances, triploidy, and regions of allelic homozygosity (which may indicate uniparental disomy or parental consanguinity). The superior diagnostic power of CMA compared with karyotyping is well established in the pediatric and prenatal literature; however, exploration of the use of this technology for studying POC samples has been somewhat limited to date.9,10,12,14,15,18,20,21,22,23,24,25,26,27,28 Specifically, CMA is a particularly attractive technology for use in the study of POC samples because it is performed using extracted cellular DNA, significantly improving the likelihood of obtaining a result. Unlike karyotype analysis, CMA can even be performed on DNA extracted from formalin-fixed and paraffin-embedded tissues (FFPE), a process that is undergone by nearly every POC sample subjected to histopathological evaluation.27 Although evaluation of aneuploidies by fluorescence in situ hybridization (FISH) analysis or quantitative PCR-based methods is possible with both fresh and FFPE samples, a significant limitation is the limited number of targets or chromosomes that may be interrogated simultaneously.8,29 Notably, SNP-based CMA can detect the presence of contaminating maternal DNA, drastically reducing the likelihood of a false-negative result due to MCC.
Although many chromosomal abnormalities that lead to a miscarriage are sporadic and have a low risk of recurrence, some abnormalities are expected to substantially increase the risk of recurrence and may necessitate parental karyotyping and/or prenatal diagnosis in future pregnancies. The American College of Obstetricians and Gynecologists, the Royal College of Obstetricians and Gynecologists, and the American Society for Reproductive Medicine all advocate chromosomal evaluation of POC samples as part of the clinical management of couples with recurrent pregnancy loss.1,30,31 By identifying the 50–60% of women whose pregnancy loss is due to a gross chromosomal abnormality, microarray analysis prevents patients from undergoing costly and unnecessary evaluations, whereas a negative result indicates that investigations targeted toward a variety of systemic disorders, including endocrinologic, hematologic, anatomic, or immunologic problems, are warranted.1,32 Importantly, identification of specific chromosome abnormalities provides valuable information that directs counseling for estimation of risk for recurrent pregnancy loss and evaluation of increased risk of having live-born offspring with congenital anomalies and/or neurocognitive disabilities. In addition, although significant weight is put on demonstrable improvements in patient outcomes and in cost savings, the psychological benefit of identifying the etiology of a fetal loss cannot be understated. Bardos et al.33 recently evaluated public attitudes in the United States regarding pregnancy loss and found not only that there were many misconceptions surrounding the frequency and causes of miscarriages but also that many women who had experienced pregnancy loss felt guilt regarding the loss and a significant sense of isolation. By offering women and their partners unambiguous answers about pregnancy loss, it is hoped that the adverse psychosocial impact of pregnancy loss can be greatly ameliorated. In this retrospective study of more than 8,000 consecutive cases, we evaluated the success rate, efficacy, and spectrum of abnormalities identified by CMA in POCs.
Materials and Methods
Samples
Sample types submitted for analysis included fresh tissue, FFPE tissue, chorionic villi, cultured cells, amniotic fluid, and extracted DNA. DNA extraction from fresh and FFPE POC tissues was performed using Maxwell 16 Tissue and Maxwell 16 FFPE Tissue LEV DNA Purification kits, respectively (Promega, Madison, WI). For FFPE blocks and slides, the area of interest containing fetal tissue was determined via examination of hematoxylin and eosin (H&E)-stained sections. Following the tissue demarcation, microdissection, and DNA extraction, sample restoration (for FFPE tissues) was performed prior to SNP array analysis. For BAC array–based comparative genomic hybridization (arrayCGH), “sample restoration” was not a step in the sample preparation.
Microarray analysis
During the period for which data were analyzed, the microarray platforms utilized evolved from BAC clone–based arrayCGH to oligonucleotide-based arrayCGH to the current SNP-based microarray. The majority of samples were evaluated using the SNP microarray (CombiSNP Array). The cytogenomics laboratory at CombiMatrix has offered and continues to offer only microarray-based testing for POC samples. With increasing resolution achieved by transitioning from BAC arrayCGH to SNP microarrays, the threshold of detection of copy-number variation also evolved to a much higher resolution.
BAC arrayCGH. Whole-genome arrayCGH was performed using the CombiBAC array for pregnancy loss, manufactured in house at CombiMatrix. This array utilized 3,520 unique large-insert clones providing an average probe spatial resolution of ~800 kb. Equivalent amounts (1 μg) of patient and male reference gDNA (Promega) were differentially labeled with Alexa Fluor 5 and Alexa Fluor 3 fluorescent nucleotides using random prime labeling protocol (Bioprime Total Genomic Labeling System; Invitrogen, Grand Island, NY), purified, coprecipitated with Cot-1 DNA, and cohybridized for 18 hours. Hybridized arrays were washed and scanned on GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA) at 10-μm resolution. DNA copy number in the patient sample was evaluated relative to reference diploid male control using DNAnalytics software (CombiMatrix, Irvine, CA), and evaluation of copy-number changes was performed across covered regions of the genome. Copy-number losses or gains were called at an effective resolution of 2.4 Mb or more (three or more contiguous BAC clones).
Oligonucleotide arrayCGH. Whole-genome arrayCGH was performed using the DNAarray for pregnancy loss analysis (a customized oligonucleotide array manufactured by Agilent Technologies, Santa Clara, CA). This array included 180,334 oligonucleotide probes covering coding and noncoding human genome sequences at an average spatial resolution of ~16 kb. Equivalent amounts (1 μg) of patient and reference male gDNA (Promega) were differentially labeled, purified, precipitated, and hybridized as described for BAC arrayCGH. Postwash arrays were scanned on the Agilent Microarray Scanner at 5-μm resolution. Copy-number change was evaluated in relation to a reference diploid sex-matched DNA sample using Nexus software (BioDiscovery, Hawthorne, CA), and targeted evaluation of copy-number changes involving more than six probes was performed in all regions of the genome. Copy alterations were called at an effective resolution of 112 kb or more (seven or more contiguous probes).
Whole-genome SNP array. Whole-genome CombiSNP array was performed using the SNP-based CytoSNP-850K array manufactured by Illumina (San Diego, CA) and using 851,622 SNP probes that provided a median resolution of 1 kb within gene-rich regions and 5 kb outside of gene-rich regions. Array processing was performed according to the manufacturer’s recommendations. DNA was evaluated for copy-number changes using Genome Studio (Illumina) and Nexus software (BioDiscovery) involving 16 or more probes for copy-number variants (CNVs) and ≥5 Mb for regions of homozygosity (ROHs). Genomic imbalances are reported using UCSC Human Genome Build 19 (NCBI build 37, February 2009). Mosaicism for partial or whole-chromosome aneuploidy is reported when present at or above the detection threshold of 15%. Maternal cell contamination was estimated from the B-allele frequency plot and, if present, at a level that interfered with interpretation of fetal results, the limitations of interpretation were indicated on the report. Correlation with microscopic evaluation of fresh tissues or H&E-stained FFPE slides was undertaken for all cases.
Reporting criteria for deletions and duplications by the three platforms were determined by multiple factors, including array resolution (~20 kb for SNP arrays to 2.4 Mb for BAC arrayCGH), size of the CNV, gene content (specifically, dosage-sensitive genes with explicit disease association), and/or overlap with a cytogenetically relevant deletion/duplication critical region. Most clinically significant CNV calls are >2 Mb.
FISH
Interphase FISH to evaluate triploidy/polyploidy was performed for all non-FFPE samples evaluated by BAC-based and oligonucleotide-based arrayCGH. FISH analysis utilized commercially available probes from Abbott Molecular specific to the satellite DNA of chromosomes 3, X and Y (Abbott Laboratories, Abbott Park, IL). A single-cell suspension was prepared from the fresh POC tissue and subsequently immobilized on slides. Probe hybridization, posthybridization washes, and signal enumeration were performed according to the manufacturer’s protocol and guidelines. A slide prepared from a normal male peripheral blood sample was used as a control.
Results
Specimen types and characteristics
Over a 44-month period, 8,298 samples were received for microarray analysis. Of these, 180 specimens were precluded from analysis as a result of client-initiated test cancellation, leaving a total of 8,118 samples that have undergone testing, analysis, review, and reporting processes. These included 6,196 (76.3%) fresh POC samples and 1,823 FFPE specimens (22.5%), and the remaining 99 samples (1.2%) were a variety of sample types, including amniotic fluid, cultured cells, DNA, and cord blood. Most of the samples (81.6%) were evaluated by the whole-genome SNP-based microarray.
Clinical ascertainment
The most common referral reason was recurrent pregnancy loss (including spontaneous abortion), comprising 6,738 (83%) cases. Intrauterine fetal demise, stillbirth, and neonatal death were the second most common referral reasons, comprising 781 (9.6%) samples received. A subset of cases (262; 3.2%) was ascertained for a variety of fetal structural abnormalities. Known, suspected, or a family history of chromosomal abnormality and a variety of additional nonspecific reasons were indicated in the remaining 337 cases (4.2%).
Maternal age and fetal gestational age
Maternal age was recorded for 8,290 cases and included 2,043 patients younger than 30 years (24.7%), 3,130 patients between 30 and 35 years (37.8%), 2,255 patients between 36 and 40 years (27%), and 862 patients older than 40 years (10.5%) ( Figure 1 ). Approximate fetal gestational age (GA) was provided only for a subset of patients (1,818; 22.4%); GA was <12 weeks for 1,055 cases (58%), between 12 and 15 weeks for 265 cases (14.6%), 16 and 20 weeks for 223 cases (12.3%), and greater than 20 weeks for 275 cases (15.1%). Although these data may not be extrapolated to the entire series, it is likely that the majority of samples were from pregnancies <20 weeks of gestation.
Successful CMA results for fresh versus FFPE samples
Overall, 7,396 samples (91.1%) were successfully processed and results were reported. Analytical success as a distribution of sample type was as follows: 92.4% for fresh tissue samples (5,725 of 6,196), 86.4% for FFPE samples (1,575 of 1,823), and 97% for other sample types (96 of 99). The most common reason for failure to report results was insufficient (or any) fetal tissue.
Overall results
Abnormal results were obtained in 53.7% of samples (3,975 of 7,396), normal results were obtained in 44.3% (3,272 of 7,396), and variants of uncertain significance (VOUS) were obtained in the remaining 2.0% (149 of 7,396) ( Tables 1 and 2 ). Cases reported as normal were identified as male in 49.6% (1,624 of 3,272) and as female in 50.4% (1,648 of 3,261). For FFPE samples, the male-to-female ratio for cases reported as normal was 0.9; this equivalent distribution of fetuses of both genders excludes the possibility of any overdiagnosis of females (false negative) due to maternal cells/tissue contributing to the analysis. In approximately 94% of cases with abnormalities, the genomic or chromosomal imbalance would be predicted to be causative of the pregnancy loss or fetal demise ( Table 2 ).
Abnormality rates in fresh versus FFPE samples
Abnormal results were observed in 48.6% of all FFPE samples analyzed (765 of 1,575). Abnormal results were identified in 55.1% of non-FFPE samples (3,210 of 5,821), the majority of which were fresh POCs ( Table 1 ).
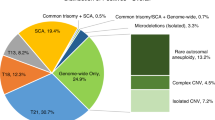
Spectrum of abnormalities
Approximately 18.4% of samples evaluated by BAC-based or oligonucleotide arrayCGH were interrogated only for segmental and whole-chromosome aneuploidies and ploidy abnormalities (by FISH). The remaining 81.6% of samples evaluated by the SNP-based array provided additional information, including whole-genome uniparental isodisomy (molar pregnancy) and copy-neutral regions of allelic homozygosity. Maternal cell contamination above a certain level (5–10%) was detectable by the SNP array analysis.
Trisomy and multiple aneuploidies
Trisomy for a single chromosome or for two or more chromosomes (multiple trisomy) was the most common abnormality; it was identified in approximately 67% of cases (2,645 of 3,975). Single-chromosome trisomy represented 95% of these cases (2,516 of 2,645), and multiple trisomy comprised the remaining 5% (129 of 2,645) ( Table 2 ) and Supplementary Figure S1 online). In 11 cases, a trisomy was identified in addition to uniparental isodisomy for a different chromosome (9 cases) or multiple regions of allelic homozygosity (2 cases). The uniparental isodisomy (isoUPD) and multiple ROHs were not likely to have contributed to the pregnancy loss. This increases the number of cases with trisomies to 2,656. With respect to single-chromosome aneuploidies, trisomy 16 was the most common and was observed in 623 cases (23.5% of all aneuploidies and 15.7% of all abnormalities), and trisomy 22 was the second most common (379 cases, 14.3% of all aneuploidies and 9.6% of all abnormalities). Trisomy 15 and trisomy 21 were equally common, at 11% and 11.3% of all aneuploidies, respectively (290 and 298 cases, respectively). Trisomy 13, trisomy 18, and trisomy 14 comprised 6.6, 4.3, and 3.3% of all aneuploidies, respectively ( Table 3 ). The combined frequency of trisomy for all other chromosomes (except chromosome 1) was 20.7% of all trisomy cases (551 cases).
Monosomy for X chromosome and autosomes
Monosomy for chromosome X was observed in 11.2% of abnormal cases (446 of 3,975). An additional 33 cases harbored a second aneuploidy in addition to monosomy X. Monosomy for autosomes was infrequent, with 26 cases identified (24 cases with monosomy 21 and one case each with monosomy 22 and monosomy 4).
Polyploidy and molar pregnancy
Triploidy was identified in 468 cases (11.8% of all abnormalities). In an additional 86 cases, aneuploidy for one or more chromosomes was observed in addition to the triploid status (hypotriploidy or hypertriploidy), indicating an overall frequency ~14% (554 of 3,975 cases) ( Table 2 ; Supplementary Figure S2 online). An XXX chromosome complement was seen in 40% of the triploid cases, XXY was seen in 58%, and XYY was seen in the remaining 2% cases. Only two cases with tetraploidy were identified by FISH analysis that was utilized for all cases evaluated by BAC or oligo arrayCGH (92,XXXX in both cases). However, the number of cases with tetraploidy may have been slightly underestimated because tetraploidy was not evaluated in FFPE samples and samples evaluated by SNP array. Whole-genome isoUPD consistent with a complete molar pregnancy was observed in 37 cases (~1% of all abnormalities) (Supplementary Figure S3 online). In two of these cases, the SNP B-allele profile was suggestive of possible dispermy. No individual confirmation was possible.
Segmental aneuploidies (deletions, duplications, and complex abnormalities)
Segmental imbalances were observed in 181 cases (4.5% of all abnormalities). These included 60 cases with deletions and 21 with duplications; in 20 cases with deletions and 14 cases with duplications, the size of the segment was >10 Mb, implying that successful high-resolution chromosome analysis may have detected the abnormality in some of these cases. In 54 of 181 cases, both a terminal deletion and a terminal duplication were observed, suggestive of an unbalanced translocation. These included 49 cases in which one or both involved segments were larger than 10 Mb. In 46 of the 181 cases, the nature of the segmental imbalances was considered complex (two or more deletions/duplications involving the same or two or more chromosomes). In 40 of these cases, at least one of the involved segments was >10 Mb (Supplementary Figure S4 online) (Supplementary Table S2 online).
Copy-neutral regions of homozygosity and uniparental disomy (N = 24)
Whole-chromosome isoUPD was observed in nine cases (involving chromosomes 2, 4, 13, 16, 17, 18, and 21), and segmental isoUPD was seen in three cases (chromosomes 6q, 7q, and 11q). In all nine cases with whole-chromosome isoUPD, trisomy for another chromosome was an associated finding and the most likely cause of the pregnancy loss. In six cases, two or more regions of homozygosity involving a single chromosome were observed, suggestive of mixed heterodisomy and isodisomy for the chromosome involved (involving chromosomes 1, 6, 7, 16, 19). Regions of allelic homozygosity involving multiple chromosomes were observed in six cases, suggestive of close parental relatedness; in two of these cases, trisomy for a single chromosome was also identified and likely the causative event ( Table 2 ). Chromosomal UPD and multiple ROHs (suggestive of identity by decent (IBD)) were considered incidental findings and not causative of a pregnancy loss. Although ROHs, isoUPD, and mixed iso/heteroUPD restricted to one chromosome are not considered causes of pregnancy loss, evaluation of these results may provide additional information for late gestational pregnancy losses with multiple congenital anomalies.
Mosaic abnormalities
Mosaicism for an abnormality was observed in 193 cases (4.9% of abnormal cases). This included mosaicism for an autosomal aneuploidy in 116 cases, mosaic sex chromosome aneuploidy in 35 cases, mosaicism for a chromosome copy number in cases of hypotriploidy or hypertriploidy in 13 cases, and mosaic segmental aneuploidies in 29 cases.
VOUS
VOUS were identified in 149 cases (2% of all cases analyzed). In 130 cases, the CNV(s) were 2 Mb or less and >2 Mb in the remaining 19 cases. Specifically, copy-number losses were observed in 67 cases and copy-number gains were observed in 79 cases. In 3 cases, both a loss and a gain were observed. In 10 cases, the CNV included or overlapped with a cytogenetically relevant region but with variable phenotypic effect and variable penetrance and or expressivity (Supplementary Table S3 online). These CNVs were not considered contributory to the miscarriage but were reported because of possible postnatal consequence and counseling for future pregnancies.
Maternal age and abnormal CMA
Maternal age at sample collection was recorded for 3,975 cases with abnormal CMA results. Maternal age was younger than 30 years for 18.5% cases of abnormal findings and older than 30 years for 81.5% of cases with abnormalities.
MCC
Maternal cell contamination was estimated for cases evaluated by SNP array using a semiquantitative method (based on genome-wide changes to the SNP B-allele profile) with an internally developed standard. This standard was developed by measuring changes to the SNP B-allele profile following increasing amounts of maternal DNA spiked in a fetal sample. Varying levels of MCC were recorded for 2,042 SNP array cases (1,603 fresh and 439 FFPE samples) (Supplementary Table S1 online). The frequency of MCC was higher in FFPE samples compared to fresh samples, particularly levels of MCC of 10% or more (Supplementary Table S1 online). Insignificant levels of MCC were recorded for 93% of 1,603 fresh and 60% of 439 FFPE samples. These results suggest that low levels of MCC are registered at high sensitivity and that the detection of segmental or whole-chromosome imbalances is not adversely impacted by levels of MCC less than 50%.
Discussion
Clinical pregnancy loss is influenced by many factors. Two frequently cited associations include increasing maternal age and previous losses.34,35 The most common etiology is fetal chromosomal abnormalities, particularly prior to 20 weeks of gestation. Although chromosomal factors in the patient or her partner (largely balanced chromosomal rearrangements) only account for approximately 2–5% of all couples experiencing recurrent pregnancy loss, approximately 50–60% of all first-trimester miscarriages are caused by a fetal chromosome abnormality. For an etiological explanation of such magnitude, we would assume that fetal chromosome analysis would be offered routinely for all pregnancy losses; however, historically, evaluating fetal chromosomes has been technically challenging. Traditional cytogenetic techniques, such as karyotyping, rely on obtaining healthy cells for culturing; however, obtaining such a sample from an early miscarriage or following an intrauterine fetal demise is challenging, and often fails to produce results. In addition, for archived FFPE samples, it is not possible to retrospectively perform karyotyping. Evaluation by FISH analysis is an attractive option for both fresh and FFPE samples; however, interrogating more than a few and common chromosome-specific loci is not technically viable. Additionally, the sensitivity for low-level mosaic abnormalities is challenging by karyotype and FISH methods. Professional society guidelines delineate the clear benefits of chromosomal analysis of fetal losses,1,30,31 including specific recommendations regarding parental studies, whether or not additional etiologies need to be explored, whether prenatal and/or preimplantation genetic testing should be offered, and what the couple’s recurrence risks are.
Reports of POC samples evaluated solely by CMA have been few and have mostly included a limited number of samples.25,26,27,28 The lack of data on CMA analysis of archived FFPE samples is even more apparent. Enhancements in SNP-based array technology provide significant advantages over traditional arrayCGH combined with our in-house sample handling expertise and have resulted in high success rates in the analysis of challenging POC samples.
Over a 44-month period, 8,118 POC samples were analyzed by CMA and results were successfully provided for 91.1% of the samples, which significantly outperforms traditional cell culture–based cytogenetic methodologies. Archived FFPE tissue samples can now be analyzed for whole-genome and segmental aneuploidy with increasing success (>85%). Additionally, implementation of SNP microarrays allowed identification of molar pregnancy, triploidy and uniparental isodisomy, and specimens with maternal cell contamination, thus enabling reporting of accurate fetal-specific results. As a provider of only microarray-based testing for miscarriage analysis, a comparative study with chromosome and/or FISH analysis was not possible.
The median age of our cohort of patients was 34 years, and the median patient age for those with abnormal results was 35 years. The median gestational age, when recorded, was 10 weeks and 4 days.
An abnormality frequency of 53.7% (with ~94% of abnormalities likely causative) is marginally above the conventionally stated 50% frequency.30,33 Whole-chromosome autosomal aneuploidies, as expected, were the most common abnormality and accounted for 67% cases, the majority of which (95%) were monochromosomal. Interestingly, monosomy X was observed at a frequency higher than previously reported (~11%).25,35 The frequency of ploidy changes, particularly hypertriploidy and hypotriploidy observed in this study (~14% overall), perhaps reflects the true frequency of this phenomenon as a causative abnormality in miscarriage. A particularly notable finding was the frequency of whole-genome uniparental isodisomy and pathogenomonic and genetic confirmation of complete molar pregnancy, comprising 1% of all abnormal samples; this frequency is probably higher because neither arrayCGH nor conventional cytogenetic analysis is capable of detecting whole-genome isoUPD. Overall, a conservative estimate is that approximately 5.6% of all cases with reportable findings (231 cases with abnormal and 149 with VOUS results out of 3,975 cases) would not be identifiable by a successful karyotype or FISH analysis. However, taking into account that 21.3% of successfully analyzed samples in this series were FFPE, and that cases with 25% or more MCC would not have been amenable to karyotyping or FISH, it is reasonable to suggest that the outcomes from microarray analysis are significantly superior to those of conventional cytogenetics.
The detection of segmental aneuploidies, in the form of a terminal deletion and a terminal duplication in 1.3% of abnormal cases is suggestive of an unbalanced translocation in the fetus, with the potential of being inherited from a balanced carrier parent. Although it is unclear whether a causative role may be attributed to many of these putative unbalanced translocations, an important role of such imbalances in recurrent pregnancy loss and a role for targeted prenatal diagnosis in future pregnancies, as well as a resource for future studies, are undeniable. Notably, 32% of cases with segmental aneuploidies (58/181) involved segments <10 Mb and would probably have been missed by conventional karyotype analysis (Supplementary Table S2 online). However, a very successful karyotype and/or extensive FISH analysis would probably have enabled identification of all aneuploidies, polyploidies, monosomies, and unbalanced rearrangements (involving segments >10 Mb) and would comprise more than 95% of the cases with abnormalities reported in this series.
Although the challenge of not being able to detect balanced rearrangements and low-level mosaicism (<10–15%) exist, the likelihood of a balanced rearrangement resulting in an adverse pregnancy outcome is expected to be rare.
The present study demonstrates the benefits of POC analysis by SNP-based microarray in terms of overcoming the challenges of culture failure and false-negative results due to MCC, the ability to identify whole-chromosome aneuploidy, triploidy, segmental imbalances below the standard resolution of karyotyping, and the ability to detect whole-genome isoUPD. The benefit of highly successful analysis of archived FFPE POC samples offers a new option for women who have experienced a pregnancy loss but did not have a fresh tissue sample analyzed at the time of the loss. This is particularly beneficial for cases of recurrent pregnancy loss, enabling analysis of archived FFPE samples for previous losses. Establishing a causative diagnosis reduces or eliminates prolonged medical evaluation and testing, thus reducing both short- and long-term health-care costs.
Disclosure
T.S., N.D., M.N.S., K.H., and C.D. are full-time employees at CombiMatrix and own stock in the company. The other authors declare no conflict of interest.
References
Practice Committee of the American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril 2012;98:1103–1111.
Ford HB, Schust DJ. Recurrent pregnancy loss: etiology, diagnosis, and therapy. Rev Obstet Gynecol 2009;2:76–83.
Simpson JL, Juneau ERM. Pregnancy loss. In: Gabe SG, Nimbly JR, Simpson JL, eds. Obstetrics: Normal and Problem Pregnancies, 6th edn. Elsevier Saunders: Philadelphia, PA, 2012.
Menasha J, Levy B, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med 2005;7:251–263.
Yusuf RZ, Naeem R. Cytogenetic abnormalities in products of conception: a relationship revisited. Am J Reprod Immunol 2004;52:88–96.
Nagaishi M, Yamamoto T, Iinuma K, Shimomura K, Berend SA, Knops J. Chromosome abnormalities identified in 347 spontaneous abortions collected in Japan. J Obstet Gynaecol Res 2004;30:237–241.
Dória S, Carvalho F, Ramalho C, et al. An efficient protocol for the detection of chromosomal abnormalities in spontaneous miscarriages or foetal deaths. Eur J Obstet Gynecol Reprod Biol 2009;147:144–150.
Shearer BM, Thorland EC, Carlson AW, Jalal SM, Ketterling RP. Reflex fluorescent in situ hybridization testing for unsuccessful product of conception cultures: a retrospective analysis of 5555 samples attempted by conventional cytogenetics and fluorescent in situ hybridization. Genet Med 2011;13:545–552.
Reddy UM, Page GP, Saade GR. The role of DNA microarrays in the evaluation of fetal death. Prenat Diagn 2012;32:371–375.
Gao J, Liu C, Yao F, et al. Array-based comparative genomic hybridization is more informative than conventional karyotyping and fluorescence in situ hybridization in the analysis of first-trimester spontaneous abortion. Mol Cytogenet 2012;5:33.
Stephenson M, Kutteh W. Evaluation and management of recurrent early pregnancy loss. Clin Obstet Gynecol 2007;50:132–145.
Reddy UM, Page GP, Saade GR, et al.; NICHD Stillbirth Collaborative Research Network. Karyotype versus microarray testing for genetic abnormalities after stillbirth. N Engl J Med 2012;367:2185–2193.
Carp H, Toder V, Aviram A, Daniely M, Mashiach S, Barkai G. Karyotype of the abortus in recurrent miscarriage. Fertil Steril 2001;75:678–682.
Zhang YX, Zhang YP, Gu Y, et al. Genetic analysis of first-trimester miscarriages with a combination of cytogenetic karyotyping, microsatellite genotyping and arrayCGH. Clin Genet 2009;75:133–140.
Raca G, Artzer A, Thorson L, et al. Array-based comparative genomic hybridization (aCGH) in the genetic evaluation of stillbirth. Am J Med Genet A 2009;149A:2437–2443.
Lomax B, Tang S, Separovic E, et al. Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 2000;66:1516–1521.
Bell KA, Van Deerlin PG, Haddad BR, Feinberg RF. Cytogenetic diagnosis of “normal 46,XX” karyotypes in spontaneous abortions frequently may be misleading. Fertil Steril 1999;71:334–341.
Robberecht C, Schuddinck V, Fryns JP, Vermeesch JR. Diagnosis of miscarriages by molecular karyotyping: benefits and pitfalls. Genet Med 2009;11:646–654.
van den Berg MM, van Maarle MC, van Wely M, Goddijn M. Genetics of early miscarriage. Biochim Biophys Acta 2012;1822:1951–1959.
Fritz B, Hallermann C, Olert J, et al. Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)—re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur J Hum Genet 2001;9:539–547.
Schaeffer AJ, Chung J, Heretis K, Wong A, Ledbetter DH, Lese Martin C. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet 2004;74:1168–1174.
Ballif BC, Kashork CD, Saleki R, et al. Detecting sex chromosome anomalies and common triploidies in products of conception by array-based comparative genomic hybridization. Prenat Diagn 2006;26:333–339.
Shimokawa O, Harada N, Miyake N, et al. Array comparative genomic hybridization analysis in first-trimester spontaneous abortions with ‘normal’ karyotypes. Am J Med Genet A 2006;140:1931–1935.
Hillman SC, Pretlove S, Coomarasamy A, et al. Additional information from array comparative genomic hybridization technology over conventional karyotyping in prenatal diagnosis: a systematic review and meta-analysis. Ultrasound Obstet Gynecol 2011;37:6–14.
Levy B, Sigurjonsson S, Pettersen B, et al. Genomic imbalance in products of conception: single-nucleotide polymorphism chromosomal microarray analysis. Obstet Gynecol 2014;124(2 Pt 1):202–209.
Wang BT, Chong TP, Boyar FZ, et al. Abnormalities in spontaneous abortions detected by G-banding and chromosomal microarray analysis (CMA) at a national reference laboratory. Mol Cytogenet 2014;7:33.
Kudesia R, Li M, Smith J, Patel A, Williams Z. Rescue karyotyping: a case series of array-based comparative genomic hybridization evaluation of archival conceptual tissue. Reprod Biol Endocrinol 2014;12:19.
Dhillon RK, Hillman SC, Morris RK, et al. Additional information from chromosomal microarray analysis (CMA) over conventional karyotyping when diagnosing chromosomal abnormalities in miscarriage: a systematic review and meta-analysis. BJOG 2014;121:11–21.
Russo R, Sessa AM, Fumo R, Gaeta S. Chromosomal anomalies in early spontaneous abortions: interphase FISH analysis on 855 FFPE first trimester abortions. Prenat Diagn 2016;36:186–191.
American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol 2013;122:1374–1377.
Regan L, Backos M, Rai R. The investigation and treatment of couples with recurrent first-trimester and second-trimester miscarriage. RCOG Green-top Guideline No. 17. April 2010. https://www.rcog.org.uk/globalassets/documents/guidelines/gtg_17.pdf.
Foyouzi N, Cedars MI, Huddleston HG. Cost-effectiveness of cytogenetic evaluation of products of conception in the patient with a second pregnancy loss. Fertil Steril 2012;98:151–155.
Bardos J, Hercz D, Friedenthal J, Missmer SA, Williams Z. A national survey on public perceptions of miscarriage. Obstet Gynecol 2015;125:1313–1320.
Whitley E, Doyle P, Roman E, De Stavola B. The effect of reproductive history on future pregnancy outcomes. Hum Reprod 1999;14:2863–2867.
Simpson, J, Carson, S. Genetic and nongenetic causes of pregnancy loss. Global Library of Women’s Medicine, 2013. http://www.glowm.com/section_view/item/318.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
(ZIP 854 kb)
Rights and permissions
About this article
Cite this article
Sahoo, T., Dzidic, N., Strecker, M. et al. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med 19, 83–89 (2017). https://doi.org/10.1038/gim.2016.69
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.69
Keywords
This article is cited by
-
Experience of copy number variation sequencing applied in spontaneous abortion
BMC Medical Genomics (2024)
-
Genetic testing for fetal loss of heterozygosity using single nucleotide polymorphism array and whole-exome sequencing
Scientific Reports (2024)
-
Reproductive outcomes in couples with recurrent pregnancy loss after embryonic chromosomal microarray analysis
Journal of Assisted Reproduction and Genetics (2024)
-
Association of TP53 rs1042522 G > C, MDM2 rs2279744 T > G, and miR-34b/c rs4938723 T > C polymorphisms with aneuploidy pregnancy susceptibility
BMC Pregnancy and Childbirth (2023)
-
Genetic analysis of chorionic villus tissues in early missed abortions
Scientific Reports (2023)