Abstract
Purpose:
The Inborn Errors of Metabolism Information System (IBEM-IS) collects data on the clinical history of inborn errors of metabolism (IBEMs). The IBEM-IS is accessible to metabolic clinics nationwide and seeks to (i) influence clinical management of affected individuals and (ii) provide information to support public health decision making.
Methods:
Thirty centers in 21 states are enrolling persons with newborn-screened conditions, collecting information on diagnosis and treatment at the time of enrollment and all subsequent visits. Prospective data are collected using electronic capture forms allowing aggregation of information regarding outcomes for individuals affected with IBEMs.
Results:
A total of 1,893 subjects have been enrolled in the IBEM-IS, and more than 540,000 individual data points have been collected. Data collection has been initiated for subjects with 41 of 46 conditions on the recommended uniform screening panel; 4 conditions have more than 100 subjects enrolled. Median follow-up time for subjects with more than one visit (n = 898) is 1.5 years (interquartile range = 2.2 years). Subjects with critical conditions are more likely to have emergency letters and sick-day plans. Mortality was exclusive to children with critical conditions.
Conclusion:
Large-scale prospective data can be collected for individuals with rare conditions, permitting enhanced decision making for clinical management and supporting decision making in public health newborn screening programs.
Genet Med 18 12, 1276–1281.
Similar content being viewed by others
Introduction
Nearly every baby born in the United States is tested soon after birth for inborn errors of metabolism (IBEMs) through newborn blood spot screening (NBS). This screening identifies rare and potentially life-threatening conditions and offers the opportunity for early intervention to improve outcomes for affected individuals. After more than 50 years of NBS, however, few screened conditions have an evidence base to justify consistent clinical practice strategies.
In current practice many interventions used to treat and manage these conditions are determined empirically, based on the experience of the clinician. Meanwhile, emerging new treatments and interventions prompt continued evolution of care practices, patients and their caregivers seek clear and consistent advice concerning effective interventions, and third-party payers and clinicians want evidence of best practices and clinical utility regarding the diagnosis, management, and long-term outcomes of these conditions. Because newborn-screened IBEMs are rare, an evidence base for decision making cannot be accumulated in a single center. Collaboration across multiple centers is necessary to systematically collect data on the management, treatment, and long-term outcomes of individuals with newborn-screened conditions. The Inborn Errors of Metabolism Collaborative (IBEMC) has overcome this barrier by creating a data-collection system that is accessible to metabolic clinics nationwide.
The IBEMC developed1 and is using a robust data-collection tool to document the clinical history of rare IBEMs detected by NBS. Our interest in initiating the Inborn Errors of Metabolism Information System (IBEM-IS) was twofold: (i) to influence the clinical management and care of affected individuals and (ii) to provide information to support decision making in public health. Here, we present an update on the progress of large-scale data collection and provide information about the character and potential utility of the information collected.
Materials and Methods
Creating the IBEM-IS
The development of a collaborative, interactive group of clinicians was essential to the data-collection efforts. Creation of this group was supported by the Region 4 Genetics Collaborative managed by the Michigan Public Health Institute (MPHI). Clinicians and public health representatives from the Region 4 states (Illinois, Indiana, Kentucky, Michigan, Minnesota, Ohio, and Wisconsin) formed a workgroup to define a set of critical elements needed for ongoing follow-up1 and initiated data collection for one condition, medium-chain acyl-CoA dehydrogenase (MCAD) deficiency. Additional disorders were added in a stepwise fashion to incorporate additional fatty acid oxidation disorders and additional categories of newborn-screened conditions (aminoacidopathies, organic acidemias, and others). The Region 4 workgroup subsequently defined condition-specific elements for all of the primary and secondary metabolic conditions identified on the recommended uniform screening panel.2 The first subject was enrolled in 2007; subjects with other conditions were enrolled as additional data elements became available. This constituted the first version of the IBEM-IS.
Simultaneous national efforts with a joint workgroup of the Newborn Screening Translational Research Network (NBSTRN) and the National Coordinating Center for the regional collaboratives brought together expert clinicians to further define common data elements for long-term follow-up. Incorporating the data elements from the Region 4 Workgroup and other expert input, common data elements and condition-specific elements were confirmed for metabolic conditions on the recommended uniform screening panel. These elements were translated into electronic data capture forms using Research Electronic Data Capture (REDCap) by the IBEMC in collaboration with the NBSTRN. REDCap is a secure, Web-based application that provides (i) an intuitive interface for validated data entry; (ii) audit trails for tracking data manipulation and export procedures; (iii) automated export procedures for seamless data downloads to common statistical packages; and (iv) procedures for importing data from external sources.3 The resulting data system is the Longitudinal Pediatric Data Resource (LPDR), a support tool that is available to researchers and provided by the NBSTRN. When REDCap forms became available, all data in the original version of the IBEM-IS was mapped to the new data elements and transferred from its original site to servers at MPHI. The IBEM-IS currently uses the LPDR data-collection forms and data dictionary.4 Data are collected using an instance of REDCap at MPHI; data are stored securely at MPHI.
Human subjects
Subjects are eligible for enrollment if they have a newborn-screened condition. Enrollment is accomplished after a diagnosis is confirmed by the enrolling center and prospective informed consent is obtained from the subjects or their legal guardians. Standardized consent materials were provided to all participating centers; support was given to any center that requested assistance with the preparation of a human subjects protection application and protocol submission. MPHI maintains records regarding institutional approvals, consent forms, and enrollment information for participating centers.
Addition of centers
The original 7 centers have increased in number to 30 centers in 21 states. Centers are added if they agree to data-use rules, agree to participate in the research activities of the collaborative, and have received human subjects protection approval from their institution. Funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, supports 13 centers; other centers are supported by their Regional Genetics Collaboratives. Additional centers continue to be engaged at their request; four centers are currently in the process of obtaining human subjects approval.
Data analysis
Data were exported from the IBEM-IS into SPSS statistical software for tabulation and analysis. Frequencies of emergency letters, sick-day plans, and 24-h on-call contact information were compared using chi-squared tests.
Results
The IBEM-IS
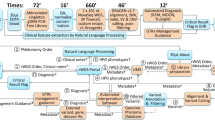
The IBEM-IS is a robust data-collection tool used to collect demographic and clinical data from patients with IBEMs and their clinicians at metabolic specialty centers in multiple states ( Figure 1 ). The data-collection instruments in the IBEM-IS are divided into three types: intake forms (n = 5), visit forms (n = 8), and special forms (n = 4). At the time a subject agrees to participate, information is collected using Web-based intake forms that include demographics, family history, health history, newborn screening assessment, and initial testing evaluation. In general, subjects are consented during a clinic visit. As a result, the intake enrollment forms and the initial visit data forms are both completed at the time of enrollment. Visit forms include information about interval health history, findings, ancillary care, laboratory studies, management, pharmacotherapy, and nutrition. Each time the enrolled individual returns to the clinic, an additional set of visit forms is completed. This allows monitoring of interval activities such as hospitalization, emergency department visits, changes in educational or other social service support, and nutritional and pharmacological interventions. Special forms are used to capture information about unique clinical interventions or circumstances such as pregnancy (1,305 elements), dialysis (93 elements), transplantation (93 elements), and study status (53 elements).
Overview of the Inborn Errors of Metabolism Information System (IBEM-IS). Clinicians, researchers, and families from several states (dark color) contribute to the IBEM-IS. Data collected using the IBEM-IS form a comprehensive record of the longitudinal progress of persons affected with newborn-screened inborn errors of metabolism.
As of 27 October 2015, 1,893 subjects have been enrolled in the IBEM-IS, which exceeded projected enrollment (n = 1,786 by March 2016). Among the 1,893 enrolled subjects, 3,850 visits are recorded in the IBEM-IS, comprising more than 540,000 individual data points ( Figure 2 ). There are 893 enrolled subjects with >1 visit, 263 with >5 visits, and 53 with >10 visits recorded. The median follow-up time recorded for subjects with more than one visit is 1.5 years (interquartile range = 2.2 years). The overall range of recorded follow-up times is 0.0–8.3 years; the longest individual follow-up is from a subject with maple syrup urine disease. These data represent a total of 1,902.7 person-years of follow-up. Of particular note, subjects with MCAD deficiency were the first to be enrolled in the IBEM-IS, and therefore they have the longest recorded per-patient follow-up. Of the subjects with MCAD deficiency, 35 have at least 5 years of follow-up data recorded ( Figure 3 ).
Follow-up time by condition. The median follow-up time, interquartile ranges, outliers, and extremes are shown in the boxes. Conditions with fewer than 10 cases are not shown. Bold dash, median; gray box, interquartile range; error bars, 95% confidence interval, open circle, outlier (value >1.5 times the interquartile range); *extreme outlier (value more than three times the interquartile range).
Demographics of enrolled subjects
The data-collection files include information from 974 males and 912 females. Age range is 22 days to 77.8 years, with an average age of 10.6 years and a median age of 7.6 years (interquartile range = 11.45 years). There are 812 subjects that are missing date of birth information. At least 198 subjects are 18 years of age or older (10.5% of the total). There are 221 families that have more than one affected child/family member enrolled. Of all cases enrolled, 1,216 (64.2%) were ascertained by NBS alone, 131 (6.9%) were ascertained by newborn screening plus at least one other diagnostic indicator, 148 (7.8%) were ascertained by clinical presentation alone, 75 (4%) were ascertained by having a family member with the condition alone, 26 (1.4%) were ascertained by having clinically identified abnormal laboratory values (independent of NBS) alone, 32 (1.7%) were ascertained by multiple diagnostic indicators not including NBS, and 265 (14%) were ascertained by an unknown or unrecorded mechanism.
Of the subjects identified, 853 (45.1%) have genetic testing results recorded and 672 (35.5%) of these have at least one pathogenic variant identified. Genetic testing was accomplished to a similar degree in three of the four largest groups of cases ascertained. Phenylketonuria (PKU) is the largest group of subjects enrolled, and 87 of 472 (18.4%) of these subjects have genetic testing recorded. MCAD deficiency, biotinidase deficiency, and classic galactosemia are the next most numerous groups, with 241 of 345 (70.7%), 91 of 198 (46%), and 97 of 181 (53.6%) of subjects, respectively, with genetic testing recorded.
With regard to ethnicity, 1,114 of 1,893 subjects have data recorded. People of European (n = 574), North American (n = 67), African (n = 24), Asian (n = 28), multiethnic (n = 159), and unknown ethnic (n = 246) descent comprise the majority of cases. With regard to race, 1,707 of 1,893 subjects have data recorded. White race (n = 1,489), black or African American race (n = 93), and multiracial (n = 44) comprise the majority of cases. A total of 100 of 1,694 subjects reported being Hispanic or Latino.
Distribution of cases by condition
Of the 46 total possible conditions for which data-collection instruments are available, 41 conditions have at least one case enrolled. Five disorders—arginase deficiency, 2,4-dienoyl CoA-reductase deficiency, medium-chain ketoacyl-CoA thiolase deficiency, medium/short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency, and 2-methyl-3-hydroxybutyric aciduria—have no cases ascertained. Nineteen conditions have 0–10 cases, 13 have 11–50 cases, 4 have 51–100, and 4 have >100 cases ( Figure 4 ). These four largest case enrollments are for galactosemia due to deficiency of galactose-1-phosphate-uridyl transferase (181 cases), biotinidase deficiency (206 cases), MCAD deficiency (348 cases), and variants of phenylalanine hydroxylase deficiency (PKU, 443 cases; hyperphenylalaninemia (H-PHE), 29 cases). Of these four largest case sets, 949 of 1,018 (93.2%) were ascertained by NBS (87.6% of galactosemia cases, 93.8% of biotinidase deficiency cases, 88.5% of MCAD cases, and 87.2% of PKU/H-PHE cases that have the mechanism of ascertainment indicated).
Using the IBEMC data to assess care for affected subjects
Emergency plan for Society for Inherited Metabolic Diseases critical disorders. The Society for Inherited Metabolic Diseases (SIMD), a professional organization representing experts in acute management of newborn screening disorders, developed a statement endorsing a subset of newborn-screened conditions as critical and requiring rapid diagnosis and immediate notification.5 We sought to learn whether providers acknowledged these different levels of acuity by providing to patients materials that reflected the potential risk of the conditions. It was anticipated that critical conditions would merit additional emphasis for urgent care, whereas noncritical ones would not likely require such interventions. There are 986 subjects with conditions designated as critical and 886 subjects with conditions that are not designated as critical. Subjects diagnosed with critical conditions have emergency letters, sick-day plans, and 24-h on-call contact information for a specialty provider given to their primary care providers more frequently than subjects diagnosed with noncritical conditions ( Table 1 ).
Survival of the IBEMC cohort. There are currently 12 deaths recorded among the 1,893 subjects enrolled in the IBEM-IS. The median age at death for these 12 subjects was 5.1 years (interquartile range = 7.5 years), with an overall range of 0.4–51.3 years. Eleven of the 12 deceased subjects had been diagnosed with SIMD critical conditions (propionic acidemia, n = 3; very-long-chain acyl-CoA dehydrogenase deficiency, n = 3; glutaric acidemia, type 1, n = 2; carnitine-palmitoyl transferase II, n = 1; galactosemia, n = 1; and maple syrup urine disease, n = 1). The one remaining deceased subject had been diagnosed with methylmalonic acidemia-cobalamin disorders. While those disorders are not formally listed as SIMD critical conditions, they often have the same presentation as methylmalonic acidemia-methylmalonyl CoA mutase, which is an SIMD critical condition.
Discussion
The IBEM-IS provides the opportunity for larger-scale analysis of newborn screening outcomes beyond the IBEM conditions encompassed by the IBEM-IS. By starting with one condition (MCAD deficiency) and using it as a paradigm for data collection, data-collection tools were successfully added for all of the metabolic conditions on the recommended uniform screening panel. In a similar fashion, by starting with a core group of clinicians and a strong infrastructure leadership, we have been successful in developing a larger group of committed clinicians who are engaged in the IBEMC. By collaborating with national experts through the NBSTRN, a support organization designed to enhance research in newborn screening, a large-scale data-collection system was developed that can be widely adapted to the long-term follow-up of newborn-screened conditions.
The initial analysis of ethnicity raised concerns that we were not ascertaining a representative group as the individuals from minority populations were seemingly underrepresented. However, after analysis of the specific conditions and their representations included in the data set, this disparity was more understandable, if not ideal. In our data set, 44% of cases have either MCAD deficiency or phenylalanine hydroxylase deficiency (H-PHE/PKU). Because these are among the more common newborn-screened conditions, and because of the strategy used for adding conditions, both are highly represented in the data set. However, phenylalanine hydroxylase deficiency (H-PHE/PKU) is rare among individuals of color,6 and MCAD deficiency is most often due to a common mutation found primarily in whites.7 In addition, Hispanic populations are poorly represented in our data set because we have not yet added centers in any of the states that have the most representation of this ethnic group (no Southwestern states, no centers in California, Texas, or Florida). Efforts to recruit centers that serve more ethnically diverse populations are ongoing.
As the IBEMC continues to evolve, it will be important to be attentive to collecting data that are as representative as possible of the population. Thus far, cases have been added to the data set as a matter of “convenience,” and no specific strategy to target any given condition has been undertaken. With time, and with consistent strategies to continue enrollment, it is anticipated that the cases will more generally reflect the incidence of these conditions among the population of individuals with newborn-screened IBEMs.
We will continue to encourage centers to enroll subjects of all ages and use all methods of ascertainment. While 71.2% of enrolled patients were ascertained by NBS, almost 30% were identified through other mechanisms. The cases ascertained using other strategies can be compared with those identified through newborn screening, which will add to the understanding of the natural history of IBEM.
For a more complete understanding of IBEM, genotyping will be an important factor in the complete analysis of outcomes. Because subjects are enrolled based on their clinical experiences, access to their genotypes has depended on (i) what was felt to be clinically indicated by their providers, (ii) what was permitted by their insurance, and (iii) the individual subject’s desire to know his or her genotype. Nonetheless, for specific conditions in the data set, genotyping has been relatively substantial. For example, 61 of 87 cases (70.1%) with very-long-chain acyl-CoA dehydrogenase deficiency have recorded genotypes that will provide a critical point of comparison for analyses about outcomes for that condition. As the price of genotyping decreases, it is anticipated that there will be more complete data for this essential data element.
The results regarding assessment of care illustrate the types of information that could be available from analysis of the data set. Our observations regarding plans for care of individuals affected with critical conditions are perhaps not surprising, but they are certainly reassuring with regard to the rigor of care provided for children with critical conditions. Individuals with conditions that should trigger immediate neonatal screening remain in need of continuing plans for emergency care; our data support the acknowledgment of this observation by members of the collaborative. In a similar fashion, the profile of mortality among involved individuals reflects the spectrum of severity expected in the screened conditions. In fact, all of the recorded deaths occurred among individuals with critical conditions, which reminds us that these are complex and medically risky conditions despite early identification and care.
Initiating, continuing, and maintaining data collection is expensive but essential to the understanding of longitudinal outcomes in newborn-screened conditions. Several additional factors will continue to present challenges in continuing this effort. Perhaps the most difficult is the sustained engagement of both subjects and collaborators necessary to maintain data collection. For some conditions, gathering sufficient numbers of cases will remain challenging and elusive. For the more “common” of the NBS conditions, a sufficient sample size for analysis is an achievable outcome, but for very rare disorders, the available sample size may allow only descriptive studies for some time to come.
Our goal is to participate in establishing best practices and clinical utility regarding the diagnosis, management, and long-term outcomes of newborn-screened IBEMs. Beginning with a single individual enrolled in 2007, we now have emerging information about the clinical course of an increasing number of individuals with rare conditions. Members of the IBEMC are encouraged by our success and hopeful that this sustained effort will yield important improvements in the clinical management of IBEMs. The IBEM-IS should permit the assessment of clinical care strategies when comparing differences between centers, allow the systematic collection of information about clinical outcomes that will lead to definition of clinical care guidelines, and provide evidence for improvements in immediate approaches to clinical care. Information from this project and its companion projects will build the evidence base required to provide optimal care management and allow affected persons to achieve their full potential for healthy and productive lives.
Disclosure
None of the authors or members of the IBEMC have any commercial association that would create a conflict of interest with the information presented in this manuscript.
References
Berry SA, Jurek AM, Anderson C, Bentler K ; Region 4 Genetics Collaborative Priority 2 Workgroup. The inborn errors of metabolism information system: A project of the Region 4 Genetics Collaborative Priority 2 Workgroup. Genet Med 2010;12(12 suppl):S215–S219.
Health Resources and Services Administration Advisory Committee on Heritable Disorders in Newborns and Children. Recommended uniform screening panel. http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/recommendedpanel/. Accessed 28 August 2015.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–381.
Newborn Screening Translational Research Network. Longitudinal Pediatric Data Resource (LPDR). https://www.nbstrn.org/research-tools/longitudinal-pediatric-data-resource. Accessed 14 September 2015.
Society for Inherited Metabolic Disorders. SIMD Position Statement: identifying abnormal newborn screens requiring immediate notification of the health care provider. 21 August 2014. http://www.simd.org/Issues/SIMD%20NBS%20Critical%20Conditions%20policy%20statement.pdf. Accessed 20 September 2015.
Hofman KJ, Steel G, Kazazian HH, Valle D. Phenylketonuria in U.S. blacks: molecular analysis of the phenylalanine hydroxylase gene. Am J Hum Genet 1991;48:791–798.
Khalid JM, Oerton J, Cortina-Borja M, et al.; UK Collaborative Study of Newborn Screening for MCADD. Ethnicity of children with homozygous c.985A>G medium-chain acyl-CoA dehydrogenase deficiency: findings from screening approximately 1.1 million newborn infants. J Med Screen 2008;15:112–117.
Acknowledgements
The research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Development, National Institutes of Health, under award number 5R01HD069039. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The Newborn Screening Translational Research Network, which is funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (HHSN275201300011C), facilitated this research. In particular, notable contributions were made by Kristi Bentler, MS, RN, CN, Amy Brower, PhD, and Jen Loutrel, MS, in the development of REDCap electronic data capture forms, creating the Long-term Pediatric Data Resource.
The members of the IBEMC are as follows: University of Colorado School of Medicine and Children’s Hospital Colorado (Janet Thomas, Melinda Dodge); Emory University Department of Human Genetics (Rani Singh, Sangeetha Lakshman, Katie Coakley, Adrya Stembridge); University of Iowa Health Care (Alvaro Serrano Russi, Emily Phillips); Ann and Robert H. Lurie Children’s Hospital of Chicago (Barbara Burton, Clare Edano, Sheela Shrestha); University of Illinois (George Hoganson, Lauren Dwyer); Indiana University (Bryan Hainline, Susan Romie, Sarah Hainline); University of Louisville (Alexander Asamoah, Kara Goodin, Cecilia Rajakaruna, Kelly Jackson); Johns Hopkins (Ada Hamosh, Hilary Vernon, Nancy Smith); University of Michigan (Ayesha Ahmad, Sue Lipinski); Wayne State University Children’s Hospital of Michigan (Gerald Feldman); University of Minnesota (Susan Berry, Sara Elsbecker); Minnesota Department of Health (Kristi Bentler); University of Missouri (Esperanza Font-Montgomery, Dawn Peck); Duke University (Loren D.M. Pena, Dwight D. Koeberl, Yong-hui Jiang, Priya S. Kishnani); University of Nebraska (William Rizzo, Machelle Dawson, Nancy Ambrose); Children’s Hospital at Montefiore (Paul Levy); New York Medical College (David Kronn); University of Rochester (Chin-to Fong, Kristin D’Aco, Theresa Hart); Women’ and Children’s Hospital of Buffalo (Richard Erbe, Melissa Samons); Cincinnati Children’s Hospital Medical Center (Nancy Leslie, Racheal Powers); Nationwide Children’s Hospital (Dennis Bartholomew, Melanie Goff); Oregon Health and Science University (Sandy vanCalcar, Joyanna Hansen); University of Pittsburgh School of Medicine (Georgianne Arnold, Jerry Vockley); Children’s Hospital of Pittsburgh of UPMC (Cate Walsh-Vockley); Medical College of Wisconsin (William Rhead, David Dimmock, Paula Engelking, Cassie Bird, Ashley Swan); University of Wisconsin (Jessica Scott Schwoerer, Sonja Henry); West Virginia University (TaraChandra Narumanchi, Marybeth Hummel, Jennie Wilkins); Sanford Children’s Specialty Clinic (Laura Davis-Keppen, Quinn Stein, Rebecca Loman); Michigan Public Health Institute (Cynthia Cameron, Mathew J. Edick, Sally J. Hiner, Kaitlin Justice, Shaohui Zhai).
Author information
Authors and Affiliations
Consortia
Corresponding author
Rights and permissions
About this article
Cite this article
Berry, S., Leslie, N., Edick, M. et al. Inborn Errors of Metabolism Collaborative: large-scale collection of data on long-term follow-up for newborn-screened conditions. Genet Med 18, 1276–1281 (2016). https://doi.org/10.1038/gim.2016.57
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.57
Keywords
This article is cited by
-
Health services use among children diagnosed with medium-chain acyl-CoA dehydrogenase deficiency through newborn screening: a cohort study in Ontario, Canada
Orphanet Journal of Rare Diseases (2019)
-
Long-term follow-up in newborn screening: the role of collaboration
Genetics in Medicine (2016)