Abstract
Purpose:
Postzygotic activating mutations of PIK3CA cause a wide range of mosaic disorders collectively referred to as PIK3CA-related overgrowth spectrum (PROS). We describe the diagnostic yield and characteristics of PIK3CA sequencing in PROS.
Methods:
We performed ultradeep next-generation sequencing (NGS) of PIK3CA in various tissues from 162 patients referred to our clinical laboratory and assessed diagnostic yield by phenotype and tissue tested.
Results:
We identified disease-causing mutations in 66.7% (108/162) of patients, with mutant allele levels as low as 1%. The diagnostic rate was higher (74%) in syndromic than in isolated cases (35.5%; P = 9.03 × 10−5). We identified 40 different mutations and found strong oncogenic mutations more frequently in patients without brain overgrowth (50.6%) than in those with brain overgrowth (15.2%; P = 0.00055). Mutant allele levels were higher in skin and overgrown tissues than in blood and buccal samples (P = 3.9 × 10−25), regardless of the phenotype.
Conclusion:
Our data demonstrate the value of ultradeep NGS for molecular diagnosis of PROS, highlight its substantial allelic heterogeneity, and confirm that optimal diagnosis requires fresh skin or surgical samples from affected regions. Our findings may be of value in guiding future recommendations for genetic testing in PROS and other mosaic conditions.
Genet Med advance online publication 02 February 2017
Similar content being viewed by others
Introduction
Several developmental disorders are caused by mutations that occur early during embryogenesis (called “postzygotic” mutations) and result in a mosaic organism composed of genetically different cell populations despite their common origin.1,2 Although various mosaic syndromes have been recognized for many years, most of their underlying genetic causes have been identified only recently, with the advent of next-generation sequencing (NGS). Following identification of a postzygotic gain-of-function mutation in AKT1 as the cause of Proteus syndrome,3 several mosaic overgrowth conditions have been associated with postzygotic mutations in other members of the PI3K-AKT-mTOR pathway.4,5,6,7,8,9 We and others reported postzygotic activating mutations of PIK3CA in a wide spectrum of developmental phenotypes previously recognized as distinct clinical entities, such as megalencephaly-capillary malformation (MCAP) syndrome,4 congenital lipomatous overgrowth, vascular malformations, epidermal nevi, scoliosis/skeletal and spinal (CLOVES) syndrome,7 and fibroadipose overgrowth (FAO).5 It was also proposed that at least some cases of Klippel-Trenaunay syndrome (KTS), a poorly defined umbrella term for disorders combining vascular malformation and mosaic limb overgrowth, may be caused by PIK3CA mutations.7,10,11 Key features of these distinct but overlapping PIK3CA-related disorders comprise congenital (or early childhood) and sporadic segmental overgrowth of varied body parts and tissues combined with vascular malformations, mosaic skin lesions such as epidermal nevi, and acral anomalies.12,13 In addition to these specific syndromes, postzygotic PIK3CA mutations were described in various isolated segmental or focal forms of overgrowth, including hemimegalencephaly (HMEG),6 megalencephaly (MEG),14 focal cortical dysplasia (FCD),15,16 hemihypertrophy, and macrodactyly.17 These genetic findings have significantly challenged the historical clinical classification of PIK3CA-related disorders. To avoid any further confusion in the field, the umbrella term of PIK3CA-related overgrowth spectrum (PROS) was proposed to designate known and emerging phenotypes caused by postzygotic PIK3CA mutations.13
PIK3CA encodes the p110a catalytic subunit of the phosphatidylinositol 3-kinase and is the most commonly mutated proto-oncogene in human somatic cancer.18 Although dozens of activating PIK3CA mutations have been reported in tumors, three mutational hot spots with strong oncogenic activity (encoding p.Glu542Lys, p.Glu545Lys, and p.His1047Arg) account for ~80% of all somatic driver mutations in PIK3CA.18,19,20 Most identified mutations in patients with PROS have been reported in multiple unique somatic cancer samples,21 and the three aforementioned cancer hot-spot mutations have been found in a majority of patients with CLOVES, KTS, FAO, HMEG, and macrodactyly.5,6,7,12,17 A broader spectrum of rare (and possibly milder) PIK3CA mutations has been reported in patients with MCAP, along with some instances of de novo germ-line mutations.4,14
Molecular diagnosis of PROS poses several challenges in terms of both biological samples and methodology. PIK3CA mutations can be found at greatly varying levels depending on the tissue tested and require sensitive methods for detecting low levels of mosaicism (<5%).4,13 Several research studies have described PIK3CA mutations in various disorders of PROS, and preliminary recommendations for clinical and molecular diagnosis have emerged from these fragmented results.13 However, our knowledge of the molecular basis of PIK3CA-related disorders remains limited, and no studies of NGS-based PIK3CA sequencing in clinical settings have been reported. Here, we describe our experience with 162 patients referred for PIK3CA testing at a clinical diagnostic laboratory, thus providing data regarding indications for testing, diagnostic yield, tissues assayed, levels of mosaicism, and spectrum of pathogenic mutations.
Materials and Methods
Study subjects
Our patient population consisted of 162 consecutive individuals with presumptive PROS referred to the molecular diagnostic laboratory of Dijon University Hospital for PIK3CA testing between October 2012 and June 2015. Referring physicians provided indication for testing as well as additional clinical information regarding brain and/or body overgrowth, vascular malformations, epidermal nevus, and acral anomalies (i.e., macrodactyly, syndactyly, polydactyly, and sandal-gap toes). Photographs of patients were also provided when available. Consistent with preliminary recommendations,13 minimum clinical criteria for testing included congenital, sporadic, and segmental overgrowth of adipose, muscle, skeletal, and/or cerebral tissue combined with at least one other key feature suggestive of PROS (“syndromic” category, Table 1 ). As previously proposed, individuals with isolated focal or segmental overgrowth were also considered to have presumptive PROS and were tested for PIK3CA mutations (“isolated” category; Table 1 ). Finally, we subdivided patients based on the presence or absence of brain overgrowth (i.e., megalencephaly or hemimegalencephaly (MEG/HMEG)). A list of all 162 affected individuals with clinical information, identified PIK3CA mutations, tissues tested, and mutant allele levels is provided in Supplementary Table S1 online. We obtained written informed consent from all patients or their legal representatives, and the ethics committee of Dijon University Hospital approved the study.
Samples
PIK3CA testing for the diagnosis of PROS usually requires specimens from an affected region, such as a skin biopsy or a surgical sample from an overgrown tissue. Previous reports have shown that postzygotic PIK3CA mutations are generally not detectable in blood samples of affected individuals, except in patients with MCAP syndrome.4,13,14 We therefore strongly encouraged referring physicians to provide freshly obtained affected tissue samples such as skin biopsy specimens or surgical specimens from debulking procedures. Blood samples were accepted as primary samples for PIK3CA screening only for individuals with MEG/HMEG. We extracted genomic DNA from fresh skin, cultured skin fibroblasts, blood, saliva, cheek swabs, and surgical specimens from overgrown regions using the Gentra Puregene Blood and Tissue Extraction Kit (Qiagen, Courtaboeuf, France). We assessed genomic DNA integrity and quantity using agarose gel electrophoresis, NanoDrop spectrophotometry, and Qubit fluorometry (Thermo Fischer Scientific, Courtaboeuf, France).
Ultradeep sequencing of PIK3CA
We screened all PIK3CA coding exons (RefSeq accession number NM_006218.2) for mutations in one sample from each affected individual (referred to as the “primary sample tested”; Supplementary Table S1 online). When available, fresh skin biopsy samples and surgical specimens were preferred over cultured skin fibroblast, buccal, and blood samples. We amplified regions of interest using custom intronic primers (Supplementary Table S2 online) and long-range polymerase chain reactions with the PrimeSTAR GXL DNA Polymerase (Takara Bio, Saint-Germain-en-Laye, France). We pooled, purified, and quantified polymerase chain reaction amplicons from each affected individual. We prepared libraries using the Nextera XT DNA Sample Preparation kit (Illumina, Paris, France), which involves simultaneous fragmentation and tagging of DNA fragments by in vitro transposition.22 We performed paired-end sequencing reactions of 150-bp reads on a MiSeq platform using 300-cycle reagent kits (v2; Illumina, Paris, France).
Sequencing data analysis
We assessed the quality of sequencing reads with FastQC (v0.11.14)23 and removed sequencing adapters and low-quality bases using Trimmomatic (v0.35).24 We aligned reads to the human genome reference sequence GRCh37/hg19 with the Burrows-Wheeler Aligner (v0.7.12).25 We performed realignment around insertions and deletions using the Genome Analysis Toolkit (GATK, v2.8-1).26 Picard27 and the GATK were used to collect quality-control, sequencing depth, and coverage metrics. As previously described,4 we identified candidate single-nucleotide variants and small insertions/deletions by recording all sites of PIK3CA coding exons and splice junctions with at least four reads not matching the reference sequence and by using a stringent base quality threshold of 30 and a mapping quality threshold of 20. Variants were then hard-filtered to achieve minimum coverage at a site of 400 reads or more, with a mutant allele fraction of at least 0.01 (i.e., 10-fold higher than the average of 0.0010 errors per targeted base of our ultradeep sequencing assay). We annotated variants with SeattleSeq Annotation28 and focused on protein-altering and splice-site changes present at a frequency less than 0.1% in the Exome Aggregation Consortium (ExAC).29 Finally, we classified the remaining PIK3CA candidate variants according to the American College of Medical Genetics and Genomics guidelines.30
Statistical analysis
Fisher’s exact tests were used to test for significant differences in numbers of molecular diagnoses and numbers of PIK3CA mutations with strong oncogenic activity between phenotype groups. Mann-Whitney U tests were used to assess statistical differences in mutant allele fractions between tissue types.
Results
Patients and samples
The study comprised a total of 162 unrelated patients (82 females and 80 males), including 38 (23.5%) adults (defined as individuals 18 years of age and older), 118 (72.8%) minors, and 6 (3.7%) fetuses (Supplementary Table S1 online). One hundred thirty-one (81%) were in the “syndromic” category and 31 (19%) were in the “isolated” category. Brain overgrowth (MEG/HMEG) was reported for 31% (50/162) of subjects, including 40 with a clinical diagnosis of MCAP syndrome ( Figure 1a ). Six patients, including four with brain overgrowth, were fetuses. Indications for testing, as provided by referring physicians, and patient counts are listed in Supplementary Table S3 online. Of note, 14 patients (including one fetus) had an initial clinical diagnosis of Proteus syndrome. However, none met published diagnostic criteria for Proteus syndrome.31 Fresh skin biopsy specimens and surgical specimens comprised 82.7% (134/162) of all primary samples tested, and cultured skin fibroblasts accounted for 6.8% (11/162) of samples. Blood and buccal specimens were the primary samples tested for 17 affected individuals ( Figure 1b ). Consistent with testing eligibility criteria, all 11 blood samples screened for PIK3CA mutations were from individuals with MEG/HMEG, including eight with a clinical diagnosis of MCAP syndrome, two with isolated HMEG, and one with isolated MEG (Supplementary Table S1 online).
Diagnostic rate of PIK3CA sequencing. (a) Total number of syndromic (dark gray) and isolated cases (light gray) by phenotype (with and without MEG/HMEG). (b) Primary tissues tested for all patients and by phenotype. (c) Molecular diagnostic rate as percentage of total cases in all patients and by phenotype. Numbers of cases with and without disease-causing PIK3CA mutations are shown in dark and light gray, respectively. *P < 0.05 for significant differences in diagnostic rates between syndromic and isolated cases. (d) Molecular diagnostic rate by primary tissue tested.
Ultradeep sequencing of PIK3CA
NGS-based ultradeep sequencing of PIK3CA achieved a mean coverage of 6424.9-fold at coding bases and splice junctions, with 100 and 98.6% of bases covered by at least 100 and 1,000 reads, respectively. All candidate PIK3CA variants identified by our first screening were confirmed by a second, independent amplification reaction and sequencing experiment in the primary sample tested, confirming the high specificity of our assay.
Molecular diagnostic yield
We identified and confirmed a pathogenic or likely pathogenic variant of PIK3CA in 66.7% (108/162) of patients ( Figure 1c ). The molecular diagnostic rate was similar between patients with (66%) and without brain overgrowth (67%). Overall, we found a significantly lower diagnostic rate in isolated cases than in syndromic cases (35.5 vs. 74%; two-tailed P = 9.03 × 10−5, Fisher exact test); this difference remained when considering only cases without brain overgrowth (33.3 vs. 74.7%; P = 0.00057). Although we observed the same tendency in the MEG/HMEG group (40 vs. 72.5%), the difference did not reach statistical significance (P = 0.070), probably due to the small sample size.
Clinical indications with the highest diagnostic yield comprised CLOVES syndrome (48/59, 81.4%), FAO (3/3, 100%), hemifacial overgrowth with or without HMEG (5/6, 83.3%), and MCAP syndrome (29/40, 72.5%) (Supplementary Table S3 online). Of interest, 78.6% (11/14) of cases with a clinical diagnosis of Proteus syndrome had a disease-causing PIK3CA mutation, further illustrating the high frequency of misdiagnosis of Proteus syndrome.32 We found disease-causing PIK3CA mutations in 46.2% (6/13) of patients with a clinical diagnosis of KTS, further supporting the idea that at least some patients with KTS should be included in the PROS.10 Indications with the lowest diagnostic yield were isolated focal or segmental forms of overgrowth with difficult access to overgrown specimens. These included isolated hemihypertrophy or overgrown legs (2/12, 16.7%), macrodactyly (2/7, 28.6%), and HMEG (1/7, 14.3%). The only molecular diagnosis of isolated HMEG reported here was from a fetal brain sample. Brain tissues were not available for the remaining six HMEG patients (Supplementary Table S1 online).
Among the 54 patients with no PIK3CA mutation, subsequent molecular investigations identified the underlying genetic cause in two. A mosaic duplication of the 11p15.5 chromosomal region was found in a patient with an overgrown leg, and a postzygotic GNAQ mutation (NM_002072.3:c.548G>A, p.Arg183Gln) was identified in a patient with an initial clinical diagnosis of KTS, which was later revised to phakomatosis pigmentovascularis (Supplementary Table S1 online).33
As anticipated, DNA derived from fresh skin and surgical specimens provided the highest diagnostic yield (92/134, 68.7%) ( Figure 2d ). The relatively high diagnostic rate in blood and buccal samples (10/17, 58.8%) is likely due to the bias in our testing eligibility criteria (see the Materials and Methods section for details).
Levels of PIK3CA mutations by sample type. (a) Swarm plot showing mutant allele fractions in samples from patients with (light gray) and without (dark gray) MEG/HMEG. Tested samples included skin and overgrown tissues (n = 99), cultured skin fibroblasts (n = 13), saliva and buccal swabs (n = 6), and blood (n = 83). (b) Box and whisker plots showing mutant allele fractions in affected (skin and overgrown tissues) and surrogate samples (buccal and blood samples).
Sample type and mosaicism
To investigate levels of mosaicism by tissue, we performed ultradeep sequencing of mutations of interest in all available samples from the 108 patients with identified PIK3CA mutations. We screened a total of 201 specimens, most of which were fresh skin or overgrown tissues (n = 99) and blood samples (n = 83) ( Figure 2a ). The mutant allele fraction for each patient and tissue is provided in Supplementary Figure S1 online. PIK3CA mutations were detected in all fresh skin and overgrown tissues tested, with a median mutant allele fraction of 0.14 (range, 0.01 to 0.47) ( Figure 2a ); 32.3 and 73.7% of these samples had a mutant allele fraction less than 0.1 and 0.2, respectively, thus highlighting the need for sensitive PIK3CA screening methods. Among the 13 cultured skin fibroblasts tested, the mutation was absent in three (23.1%), suggesting that cultured samples may not be ideal for PIK3CA mutation testing. As expected, we found that mutant allele levels were significantly higher in fresh skin and overgrown tissues than in blood and buccal samples when considering all patients (two-tailed P = 3.9 × 10−25, Mann-Whitney U test); this observation was also true for cases without (P = 3.0 × 10−24) and with MEG/HMEG (P = 5.6 × 10−5) ( Figure 2b ). Consistent with this observation, mutant allele levels were found to be systematically lower in blood and buccal samples than in fresh skin and overgrown tissues for any given patient, regardless of the phenotype (Supplementary Figure S1 online). Mutant allele levels were significantly higher in blood and buccal samples of patients with than in those without MEG/HMEG (P = 1.5 × 10−7) ( Figure 2b ), further supporting previous observations that PIK3CA mutations are generally not detectable in blood or buccal samples of individuals with PROS, except in patients with MCAP syndrome.4,13,14 We found no such difference in mutant allele levels for skin and overgrown tissues between patients with and those without brain overgrowth (P = 0.070). We detected disease-causing PIK3CA mutations in 8.3% (5/60) of blood or buccal samples from individuals with no brain overgrowth. Mutant allele levels were generally low, with the highest peaking at 11.5% of total alleles. By comparison, PIK3CA mutations were detected in 58.6% (17/29) of blood or buccal samples from patients with MEG/HMEG, with mutant allele levels ranging from 1.4 to 50%. The only mutation present in 50% of alleles (NM_006218.2:c.3074C>A, p.Thr1025Asn) was presumed to be germ-line and confirmed as de novo given its absence in parental blood samples. The patient (P1931) had isolated MEG with no signs of body overgrowth, vascular malformation, skin mosaicism, or acral anomalies.
Distribution and oncogenic activity of PIK3CA mutations
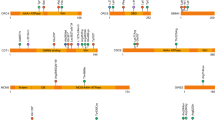
Overall, we identified 40 distinct nucleotide changes leading to 39 different amino acid changes in 108 patients ( Table 2 ). All but one mutation were postzygotic. Twenty-two (55%) were seen in more than one patient ( Table 2 and Figure 3a ). Although most mutations were missense changes, we identified two in-frame insertions/deletions encoding p.Gly106_Arg108delinsIle and p.Glu110del in four patients. Fifteen mutations (38.5%) had not been previously identified in patients with PROS, but most (37/40, 92.5%) were present in the Catalogue of Somatic Mutations in Cancer (COSMIC) at varying frequencies.21 Three (encoding p.Glu545Lys, p.His1047Arg, and p.His1047Leu) were present in ExAC,29 with each being reported in only one of ~120,000 alleles ( Table 2 ). It is unknown whether these ExAC variants are true (yet extremely rare) variants in the general population, false positives, or somatic mutations from cancer samples or patients. Of note, the only AKT1 mutation known to cause Proteus syndrome (NM_005163.2:c.49G>A, p.Glu17Lys)3 is also reported in one ExAC allele, despite being thought to be incompatible with life in a germ-line state.
Distribution and oncogenic activity of PIK3CA mutations. (a) Distribution of the mutations identified in cases without (dark gray) and with MEG/HMEG (light gray). (b) Oncogenic activity of the identified PIK3CA mutations in cases without (left pane) and with MEG/HMEG (right pane). Oncogenic activity, from weak to strong effect, is illustrated by a gray scale. Mutations with unknown effects are shown in light gray.
Among the 39 amino acid changes identified in the present study, 17 were found only in patients without MEG/HMEG, 9 were found in patients with MEG/HMEG, and 13 were shared by both groups ( Figure 3a ). To explore genotype–phenotype correlations based on the mutation effect, we used results from a recent study that aimed to characterize the oncogenic potential of somatic PIK3CA mutations found in cancer. The authors performed in vitro and in vivo cell growth and oncogenic transformation experiments to characterize the functional impact of 27 PIK3CA mutations.34 Oncogenic activity, classified from weak to strong, was available for 13 of the PIK3CA mutations identified in the present study, including the three strong oncogenic hot spots p.Glu542Lys, p.Glu545Lys, and p.His1047Arg ( Table 2 ). Consistent with previous findings,4,14,35 we found strong oncogenic mutations more frequently in patients without (50.6%) than in those with brain overgrowth (15.2%; two-tailed P = 0.00055, Fisher exact test) ( Figure 3b ). Nevertheless, only 33.3% of our patients without brain overgrowth had one of the three PIK3CA hot-spot mutations, which is in striking contrast with previous reports that such mutations were the cause of CLOVES, KTS, FAO, and other focal forms of body overgrowth in a majority of patients.12,14 Interestingly, among the five patients with MEG/HMEG caused by one of the three cancer hot-spot mutations, two were fetuses with severe brain overgrowth, two had severe hemifacial overgrowth and HMEG, and only one had a postnatal clinical diagnosis of MCAP syndrome, further supporting the idea that MCAP is rarely caused by strong oncogenic PIK3CA hot-spot mutations.14
Discussion
To our knowledge, this is the first report of the use of NGS for molecular diagnosis of disorders of the PROS in a clinical setting. Ultradeep sequencing of PIK3CA had a diagnostic rate of 66.7% in our series, with a significantly better yield for syndromic than for isolated forms. We showed that freshly obtained skin biopsy specimens and other surgical specimens from affected regions are the most suitable samples for optimal molecular diagnosis of PROS, regardless of the phenotype. Our findings also highlight the substantial allelic heterogeneity of PROS, illustrate the absence of strong genotype–phenotype correlations between clinical subgroups, underline the limitations of the historical sub-classification of these disorders into distinct syndromes, and confirm the relevance and utility of the preliminary recommendations for clinical assessment and molecular diagnosis of PROS.13
Identifying postzygotic mutations commonly present at levels below the detection threshold of capillary-based Sanger sequencing poses significant challenges to conventional molecular diagnosis. We showed that extremely deep (>1,000-fold) NGS-based sequencing of PIK3CA can readily detect levels of mutations as low as 1%. In addition to detecting single-nucleotide changes, our approach was able to identify postzygotic insertions and deletions, including a complex indel present in 7.8% of alleles and characterized by a deletion of eight and an insertion of two nucleotides. The substantial number of different disease-causing mutations (40 identified in this study alone, including 15 previously unreported) entails sequencing all coding exons of PIK3CA. Given the well-known limitations of Sanger sequencing for detecting mosaic variants,4 NGS-based ultradeep sequencing of PIK3CA appears to be the best available method for molecular diagnosis of PROS.
We observed a significantly higher number of molecular diagnoses in syndromic than in isolated forms of presumptive PROS. One reason could be that collecting affected specimens from patients with isolated anomalies may prove challenging, especially if no debulking or epilepsy surgery is planned. Our findings indicate that skin biopsy specimens from nonlesional regions and other surrogate samples are of little help for molecular diagnosis of isolated forms of PROS and emphasize the need for affected samples. Another reason could be that some of these phenotypes may be less specific than syndromic forms and due to other genetic causes, such as mutations in other members of the PI3K-AKT-mTOR pathway or mosaic forms of autosomal-dominant overgrowth syndromes, as illustrated by our patient with a mosaic duplication of the 11p15.5 region, which is a rare cause of Beckwith-Wiedemann syndrome.36
Considerations regarding which tissues to test are crucial for the optimal molecular diagnosis of mosaic disorders. Our data show that freshly obtained skin biopsy specimens and other surgical specimens from affected regions provide the best results in terms of diagnostic yield and mutant allele levels. Standard procedures of clinical laboratories for skin biopsies often involve isolating and culturing skin fibroblasts. Although based on a limited number of samples, our data suggest that fresh skin samples should be preferred over cultured cells for PIK3CA mutation screening. In addition to missing mutations that might have been detected in the original skin biopsy, cultured cells are prone to genetic alterations and drift. This may lead to false-positive results (i.e., cell culture artifacts), especially with methods meant to detect very low mutant allele levels.37
We confirmed previous observations that PIK3CA mutations are detectable in blood or buccal samples of a significant fraction of patients with MCAP syndrome.4,14 Nonetheless, our data show that blood and buccal samples lead to lower diagnostic rates and mutant allele levels than skin and other affected tissues. We therefore recommend a uniform approach based on affected skin and overgrown tissues as primary samples for molecular diagnosis of all disorders of PROS.
It has been proposed that disorders of PROS with no brain overgrowth such as CLOVES, FAO, or KTS are mostly caused by PIK3CA cancer hot-spot mutations, whereas MCAP is usually associated with rarer and milder gain-of-function mutations.14 Although we found a higher number of cancer hot-spot mutations in patients without brain overgrowth, the majority were caused by rare PIK3CA mutations, thus demonstrating that both MCAP and non-MCAP phenotypes have a broad mutational spectrum. We also found various mutations in more than one distinct clinical entity, further strengthening the idea that PIK3CA-associated phenotypes should be considered as a single spectrum. Our data confirm the advantage of using the umbrella term of PROS over historical clinical classifications for assessment of overgrowth and molecular diagnostic testing.13
In conclusion, our findings may help to define future best practices for molecular diagnosis of PROS in particular and mosaic developmental disorders in general. Along with the emergence of specific inhibitors of the PI3K-AKT-mTOR pathway,38 accurate molecular diagnoses of PROS will be crucial for paving the way toward effective treatments for patients with these disorders.
Disclosure
The authors declare no conflict of interest.
References
Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet 2013;14:307–320.
Happle R. Mosaicism in Human Skin: Understanding Nevi, Nevoid Skin Disorders, and Cutaneous Neoplasia. Springer: Heidelberg, Germany, 2014.
Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med 2011;365:611–619.
Rivière JB, Mirzaa GM, O’Roak BJ, et al.; Finding of Rare Disease Genes (FORGE) Canada Consortium. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 2012;44:934–940.
Lindhurst MJ, Parker VE, Payne F, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet 2012;44:928–933.
Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 2012;44:941–945.
Kurek KC, Luks VL, Ayturk UM, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet 2012;90:1108–1115.
Hussain K, Challis B, Rocha N, et al. An activating mutation of AKT2 and human hypoglycemia. Science 2011;334:474.
Lim JS, Kim WI, Kang HC, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med 2015;21:395–400.
Vahidnezhad H, Youssefian L, Uitto J. Klippel-Trenaunay syndrome belongs to the PIK3CA-related overgrowth spectrum (PROS). Exp Dermatol 2016;25:17–19.
Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr 2015;166:1048–54.e1.
Keppler-Noreuil KM, Sapp JC, Lindhurst MJ, et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am J Med Genet A 2014;164A:1713–1733.
Keppler-Noreuil KM, Rios JJ, Parker VE, et al. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet A 2015;167A:287–295.
Mirzaa G, Timms AE, Conti V, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight 2016;1:e87623.
D’Gama AM, Geng Y, Couto JA, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol 2015;77:720–725.
Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 2015;138(Pt 6):1613–1628.
Rios JJ, Paria N, Burns DK, et al. Somatic gain-of-function mutations in PIK3CA in patients with macrodactyly. Hum Mol Genet 2013;22:444–451.
Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–339.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70.
Hoadley KA, Yau C, Wolf DM, et al.; Cancer Genome Atlas Research Network. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929–944.
Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43(Database issue):D805–D811.
Caruccio N. Preparation of next-generation sequencing libraries using Nextera™ technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol 2011;733:241–255.
Babraham Bioinformatics. FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–595.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Broad Institute. Picard. https://broadinstitute.github.io/picard/.
SeattleSeq. SeattleSeq Annotation 138. http://snp.gs.washington.edu/SeattleSeqAnnotation138/.
Lek M, Karczewski KJ, Minikel EV, et al.; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Biesecker LG, Happle R, Mulliken JB, et al. Proteus syndrome: diagnostic criteria, differential diagnosis, and patient evaluation. Am J Med Genet 1999;84:389–395.
Cohen MM Jr, Turner JT, Biesecker LG. Proteus syndrome: misdiagnosis with PTEN mutations. Am J Med Genet A 2003;122A:323–324.
Thomas AC, Zeng Z, Rivière JB, et al. Mosaic activating mutations in GNA11 and GNAQ are associated with phakomatosis pigmentovascularis and extensive dermal melanocytosis. J Invest Dermatol 2016;136:770–778.
Dogruluk T, Tsang YH, Espitia M, et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res 2015;75:5341–5354.
Mirzaa GM, Rivière JB, Dobyns WB. Megalencephaly syndromes and activating mutations in the PI3K-AKT pathway: MPPH and MCAP. Am J Med Genet C Semin Med Genet 2013;163C:122–130.
Shuman C, Beckwith JB, Weksberg R. Beckwith-Wiedemann syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al. (eds). GeneReviews. University of Washington: Seattle, WA, 1993.
Carlson CA, Kas A, Kirkwood R, et al. Decoding cell lineage from acquired mutations using arbitrary deep sequencing. Nat Methods 2011;9:78–80.
Parker VE, Knox RG, Zhang Q, Wakelam MJ, Semple RK. Phosphoinositide 3-kinase-related overgrowth: cellular phenotype and future therapeutic options. Lancet 2015;385(suppl 1):S77.
Acknowledgements
We thank the subjects and families involved in the study, their referring physicians, and M-CM France (http://m-cmfrance.com/). We also thank the University of Burgundy Centre de Calcul (CCuB) for technical support and management of the informatics platform. This work was funded by the Agence Nationale de la Recherche (ANR-13-PDOC-0029 to J.-B.R.), the Groupe Interrégional de Recherche Clinique et d’Innovation (GIRCI) Est (to J.-B.R.), the Programme Hospitalier de Recherche Clinique (PHRC) National, and the Société Française de Dermatologie (to P.V.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figure S1
(PDF 240 kb)
Supplementary Table S1
(XLSX 31 kb)
Supplementary Table S2
(DOCX 14 kb)
Supplementary Table S3
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Kuentz, P., St-Onge, J., Duffourd, Y. et al. Molecular diagnosis of PIK3CA-related overgrowth spectrum (PROS) in 162 patients and recommendations for genetic testing. Genet Med 19, 989–997 (2017). https://doi.org/10.1038/gim.2016.220
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.220
Keywords
This article is cited by
-
Somatic mutation spectrum of a Chinese cohort of pediatrics with vascular malformations
Orphanet Journal of Rare Diseases (2023)
-
An expanded clinical spectrum of hypoinsulinaemic hypoketotic hypoglycaemia
Orphanet Journal of Rare Diseases (2023)
-
Targeted next-generation sequencing for detection of PIK3CA mutations in archival tissues from patients with Klippel–Trenaunay syndrome in an Asian population
Orphanet Journal of Rare Diseases (2023)
-
Delineation of the phenotypes and genotypes of facial infiltrating lipomatosis associated with PIK3CA mutations
Orphanet Journal of Rare Diseases (2023)
-
Epidemiology of the disorders of the Pik3ca-related overgrowth spectrum (Pros)
European Journal of Human Genetics (2023)