Abstract
Introduction:
RASopathies include disorders generally characterized by developmental delay, specific heart defects, short stature, cardiac hypertrophy, and facial dysmorphisms. Next-generation sequencing (NGS)-based panels have widespread acceptance as a diagnostic tool for RASopathies.
Materials and methods:
The first 126 patients evaluated by clinical examination and the NGS RASopathy panel at the Children’s Hospital of Philadelphia were enrolled. We calculated diagnosis rate, correlated reported clinical findings with positive or negative test results, and identified final molecular diagnoses in 28/96 patients who tested negative for RASopathies.
Results:
Twenty-four patients had pathogenic variants on the RASopathy panel, for a diagnostic yield of 19%. Reported features of pulmonic stenosis and ptosis were significantly correlated with a positive test result; no reported features were significantly correlated with a negative test result. We identified 27 different alternative diagnoses for patients originally suspected of having RASopathies.
Discussion:
This study provides information that can assist in guiding differential diagnosis and genetic testing for patients suspected of having a RASopathy disorder.
Genet Med advance online publication 20 October 2016
Similar content being viewed by others
Main
Noonan syndrome, cardiofaciocutaneous syndrome, Noonan syndrome with multiple lentigines (NSML) syndrome, and Costello syndrome share significantly overlapping genotypes and phenotypes, and are collectively called RASopathies. All are caused by dysregulation of the RAS/mitogen activation protein kinase (MAPK) pathway. Pathogenic variants in more than a dozen genes involved in the RAS/MAPK pathway have been known to cause RASopathies, and approximately 30% of patients with a clinical diagnosis were negative for all known causative genes. In Noonan syndrome, mutations in PTPN11 are the most frequent (approximately 50%), with SOS1 (10–15%), RAF1 (5–15%), KRAS (<5%), NRAS (<1%), BRAF (<1%), SHOC2 (<1%), SPRED1 (<1%), and MAP2K1 (<1%) mutations also reported.1 Recently, mutations in RIT1, SOS2, and PPPC1B have also been described as causes of human RASopathies, but the remainder of contributing genes are unknown.2,3,4 Mutations in PTPN11 and RAF occur in approximately 95% of patients with NSML syndrome; mutations in BRAF (75–80%), MAP2K1 and MAP2K2 (10–15%), and KRAS (<5%) are associated with cardiofaciocutaneous syndrome; and Noonan syndrome–like disorder with or without juvenile myelomonocytic leukemia is associated with mutations in CBL.1,5
Although there is not an absolute consensus on what phenotypes fall into the category of RASopathies, the disorders share several phenotypic features. The major clinical features of RASopathies are short stature, cardiovascular abnormalities, developmental delays, and characteristic facial anomalies. Skeletal, hematologic, and cutaneous findings can also be associated with RASopathies. This variety of presentations means that clinically diagnosing RASopathies can be challenging. In contrast to a sequential Sanger sequencing approach, a targeted next-generation sequencing (NGS) panel focused on a set of genes associated with RASopathies is a suitable, cost-effective alternative to use in the diagnosis of a RASopathy.
We conducted a retrospective review of 126 consecutive patients referred for RASopathy next-generation sequence panel testing at the Genomic Diagnostics Laboratory at the Children’s Hospital of Philadelphia. Almost all the patients (125/126) were clinically evaluated within our institution. When available, detailed electronic medical records, the patients’ clinical histories, and molecular findings were reviewed. We present and summarize the diagnostic rates, molecular findings, and reported clinical features associated with positive and negative results. In addition, we report the diagnoses in 28/96 patients who were negative for this RASopathy NGS panel. These results provide valuable data suggesting a framework for a diagnosis strategy for patients suspected of having RASopathies in the clinical and laboratory settings.
Materials and Methods
Initially, 36 patients underwent testing on 11 genes: BRAF, CBL, HRAS, KRAS, MAP2K1, MAP2K2, NRAS, PTPN11, RAF1, SHOC2, and SOS. RainDance technology (Billerica, MA) was used for enrichment, and sequencing was performed on a SOLiD 5500xl sequencer from Applied Biosystems (Foster City, CA). The remaining patients underwent testing on 12 genes (including SPRED1) with the SureSelectXT Target Enrichment System from Agilent (Wilmington, DE). MiSeq sequencing was performed following the instructions from Illumina (San Diego, CA). Read alignment and variant calling were performed with an in-house bioinformatics pipeline that incorporates Novoalign for read alignment (Selangor, Malaysia), Picard (Broad Institute, Cambridge, MA) for marking duplicates, and the Genome Analysis Toolkit (Broad Institute) for variant calling (reference sequence: hg19 Grch37 build) and variant read depth filtering (≥5×). Filtration was performed to prioritize variants in the Human Genome Mutation Database (HGMD) and/or rare variants with a coding effect such as nonsynonymous, stop loss, stop gain, start loss, indels, frameshifts, and variants within the consensus splice site (±6 bases in intron) within the RASopathy genes. Low-coverage exons (per-base coverage ≥30×) were sequenced using the dideoxy method of sequencing.
For the purpose of this study, sequence variants were classified into five categories according to the guidelines of the American College of Medical Genetics and Genomics: pathogenic, likely pathogenic, variant of uncertain significance (VOUS), likely benign, or benign.6 For clinical correlation, patients were classified as “negative” for RASopathy if they met any of the following criteria: (i) no reported variant, (ii) only a benign or likely benign variant, or (iii) reported VOUS in RASopathy panel but with a confirmed alternative diagnosis. Patients with only a VOUS and no alternative diagnosis were considered “indeterminate,” and patients with a pathogenic or likely pathogenic variant were considered “positive.” The presence of any potentially disease-causing variant was confirmed by Sanger sequence analysis.
The electronic medical records of 126 consecutive unrelated patients who had a postnatal RASopathy panel completed at the Children’s Hospital of Philadelphia were accessed with institutional review board approval. The diagnoses of these patients were identified through International Classification of Diseases, ninth revision (ICD-9), codes and careful manual review of clinical notes of all encounters at the Children’s Hospital of Philadelphia clinical network (outpatient and inpatient) by a single clinical geneticist (E.J.B.). All ICD-9 codes were manually reviewed; codes for trauma or incidental illnesses were not included. The same ICD-9 code/diagnosis was often noted at multiple encounters, and there was redundancy in some of the documentation. For example, an ICD-9 code may be for all congenital heart diseases but the clinical note specifies an atrial septal defect. To decrease clerical errors if a discrepancy was identified between ICD-9 codes and clinical notes, resolution was achieved through manual review of the clinical notes.
After collecting all the available diagnoses, correcting errors, and removing the redundancies, the curated list of diagnoses was used for statistical analysis. Phenotype categories were created based on the number of observations in the patient cohort and their relationship to RASopathies. These clinical categories were designed to be as informative as possible by balancing the specificity of the clinical diagnosis with the number of times each diagnosis is found in our entire cohort. Each category had inclusion and exclusion criteria that were uniformly applied across all patients. For example, congenital cardiac defects, which were defined as structural heart defects present at birth, did not include congenital cardiac variants such as patent foramen ovale or patent ductus arteriosus. Positive molecular or cytogenetic diagnoses with a reference laboratory variant classification of pathogenic, likely pathogenic, or VOUS were recorded. For four individuals, appropriate molecular testing had not been performed but the patients met the clinical criteria for the diagnoses. Two-sided P values and odds ratios were calculated to evaluate the percentage of specific reported findings in the mutation-positive compared to mutation-negative patients using Fisher’s exact test, with significance at P ≤ 0.05.
Results
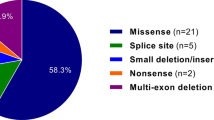
We analyzed data from 126 postnatal patients who were clinically evaluated at the Children’s Hospital of Philadelphia and who had a RASopathy panel performed between July 2013 and December 2014. A total of 10,700 ICD-9 codes were present in the charts of these 126 patients, for an average of 84.9 ICD-9 codes per patient. The RASopathy panel testing identified pathogenic variants in 24 patients: 71% (17) in PTPN11, 13% (3) in SOS1, 8% (2) in MAP2K, and 8% (2) in RAF1 (Supplementary Table S1 online). One or more VOUSs were found in nine patients, and in 95 patients either a benign variant, a likely benign variant, or no variants were found. The diagnostic yield in our population was 19% (24/126). A somatic variant in the PTPN11 gene (likely pathogenic) was identified in the bone marrow, but not buccal swabs, of a patient with acute myeloid leukemia; therefore, this patient was not included on the RASopathy-positive list.
The clinical finding that was most often reported in patients with a pathogenic RASopathy variant was pulmonic stenosis (15/24 patients; 63%), whereas developmental delay/hypotonia was the most common clinical finding reported in pathogenic variant-negative patients (41/96 patients; 43%) ( Table 1 ). The reported clinical findings that were significantly associated with a positive molecular result were pulmonary stenosis or associated anomaly (P = 0.00014) and ptosis (P = 0.016). No reported clinical findings were significantly associated with a negative molecular result. The other categories of reported findings—cardiomegaly/cardiomyopathy, developmental delay/hypotonia, short stature/failure to thrive, other congenital structural cardiac defect, hearing loss, malignancy, hydrops/edema, skin abnormality, and structural brain defects—did not show a significant difference between patients with and without a pathogenic variant.
Of the patients without a pathogenic variant, 28 received a subsequent diagnosis of a non-RASopathy disorder. These diagnoses covered a range of etiologies, including molecularly diagnosed single-gene disorders (12 patients), cytogenetic abnormalities (10 patients), clinically diagnosed syndromes (four patients), and two disease-candidate genes identified through whole-exome sequencing ( Table 2 and Supplementary Table S1 online). Neurofibromatosis type I, discovered in two patients, was the only diagnosis found in more than one patient.
Discussion
We investigated the clinical and molecular findings for 126 consecutive patients sent for RASopathy NGS, of whom 19% had a pathogenic variant (24/126) and 6% (9/126) had a VOUS. In 27% (28/102) of patients with a negative or an inconclusive test, an alternative diagnosis had been identified. We compared our gene-specific frequencies with the reported frequencies for patients referred for a variety of RASopathy-associated signs and symptoms and found them to be largely consistent.1
It has been previously reported that approximately 70% of patients with a clinical diagnosis of a RASopathy have a recognizable pathogenic mutation with molecular testing.7 We propose several hypotheses to explain the 19% detection rate in this study. The largest effect on the lower detection rate in our cohort is probably due to a broad phenotype spectrum in the patients, who may not meet clinical criteria for a RASopathy. The 28 patients with a confirmed alternative molecular diagnosis further support this hypothesis. Some of the patients appear to have limited phenotypic overlap with classic RASopathy clinical findings, and it is possible that for some patients the RASopathy testing was ordered in error. Furthermore, testing was ordered in a broad variety of settings within a teaching hospital and associated clinics, with a variety of ordering provider specialties contributing to the decision to order testing. In addition, the technical limitations of NGS in detecting variants (copy number, low coverage, and deep intronic variants) may impact the sensitivity of this test. However, the majority of reported pathogenic variants in the genes tested here are single-nucleotide variants, which would probably be identified through this test because it has 100% gene coverage of all coding regions. There may also be patients with mutations in additional RASopathy genes (RIT1, SOS2, PPPC1B) that were reported after the design of this assay, or in NF1, which was not included in the original design of this panel. Since the completion of this study, our NGS panel will expand to include NF1, RIT1, SOS2, and PPPC1B.
Using the reported clinical data, we identified clinical features that were the most predictive of a positive or a negative panel result. The reported findings that were significantly correlated with a pathogenic variant on the RASopathy panel were pulmonary stenosis and ptosis. These two diagnoses are included in the diagnostic criteria for Noonan syndrome proposed by van der Burgt, which use pulmonary valve stenosis and/or hypertrophic cardiomyopathy and typical face (including ptosis) as two of the four major named clinical diagnostic features.8 With respect to the other features, typical chest wall was not highly reported in our patients, but it might not have been noted in the medical record because it is often clinically insignificant and short stature/failure to thrive is a very common finding for a wide variety of syndromes. Although there was no significant difference between the ages of the RASopathy-positive and -negative patients, the average age of the positive patients (5.0 years) was slightly higher than that of the negative patients (2.9 years). It may be beneficial to monitor the growth and development of patients suspected of having a RASopathy to better inform testing decisions.
Among the RASopathy-negative patients with an alternative diagnosis, only one syndrome was present in more than one patient: neurofibromatosis type I (NFI). NFI was diagnosed in two of our patients and is often included in the RASopathy category of syndromes.9 Neurofibromatosis–Noonan syndrome (MIM 601321) has been recognized as a clinical condition with overlapping features of NFI and NS, and inclusion of NF1 for genetic testing in patients with clinically suspected NS has been recommended.10 Each of the other final diagnoses was found in only one patient and did not show a general predisposition for a family of disorders.
Although this study was performed in a small number of patients in a pediatric hospital setting, the results suggest that a tiered testing approach could be an option for patients with one or more features suggestive of a RASopathy disorder. A targeted NGS RASopathy panel may be an appropriate first-tier test. In individuals for whom a RASopathy panel is not informative, either exome sequencing or whole-genome array may be considered as the next option.
In summary, targeted panel testing is an efficient tool that can be used as a first-tier test in patients with a clinical diagnosis of a RASopathy disorder. For patients with negative RASopathy testing results, a review of the medical records indicated 28 patients with alternative diagnoses that span both single-gene disorders and chromosomal abnormalities. These results highlight the inherent complexities in phenotype-driven genetic testing in the pediatric population and support the application of alternative genetic testing methodologies in patients suspected of having a RASopathy disorder.
Disclosure
A.B.S: Scientific consultant Invitae, receives royalty from Agilent, honoraria from Arcadia University, Cambridge Healthtech Institute. The other authors declare no conflict of interest.
References
Romano AA, Allanson JE, Dahlgren J, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics 2010;126:746–759.
Aoki Y, Niihori T, Banjo T, et al. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am J Hum Genet 2013;93:173–180.
Cordeddu V, Yin JC, Gunnarsson C, et al. Activating Mutations Affecting the Dbl Homology Domain of SOS2 Cause Noonan Syndrome. Hum Mutat 2015;36:1080–1087.
Gripp KW, Aldinger KA, Bennett JT, et al. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am J Med Genet A 2016;170:2237–2247.
Martinelli S, De Luca A, Stellacci E, et al. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet 2010;87:250–257.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Chen PC, Yin J, Yu HW, et al. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc Natl Acad Sci USA 2014;111:11473–11478.
van der Burgt I. Noonan syndrome. Orphanet J Rare Dis 2007;2:4.
De Luca A, Bottillo I, Sarkozy A, et al. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet 2005;77:1092–1101.
Ekvall S, Sjörs K, Jonzon A, Vihinen M, Annerén G, Bondeson ML. Novel association of neurofibromatosis type I–causing mutations in families with neurofibromatosis-Noonan syndrome. Am J Med Genet A 2014;164A:579–587.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1
(XLS 22 kb)
Rights and permissions
About this article
Cite this article
Bhoj, E., Yu, Z., Guan, Q. et al. Phenotypic predictors and final diagnoses in patients referred for RASopathy testing by targeted next-generation sequencing. Genet Med 19, 715–718 (2017). https://doi.org/10.1038/gim.2016.169
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.169
Keywords
This article is cited by
-
Prenatal diagnosis of Noonan syndrome in a set of monozygotic twins- a case report
BMC Pregnancy and Childbirth (2023)
-
The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms
Leukemia (2022)
-
Variants of SOS2 are a rare cause of Noonan syndrome with particular predisposition for lymphatic complications
European Journal of Human Genetics (2021)
-
Cutaneous neurofibromas in the genomics era: current understanding and open questions
British Journal of Cancer (2018)