Abstract
Purpose:
Electronic health records (EHRs) and their associated decision support tools are potentially important means of disseminating a patient’s pharmacogenomic profile to his or her health-care providers. We sought to create a proof-of-concept decision support alert system generated from pharmacogenomic incidental findings from exome sequencing.
Methods:
A pipeline for alerts from exome sequencing tests was created for patients in the New EXome Technology in (NEXT) Medicine study at the University of Washington. Decision support rules using discrete, machine-readable incidental finding results were programmed into a commercial EHR rules engine. An evaluation plan to monitor the alerts in real medical interactions was established.
Results:
Alerts were created for 48 actionable pharmacogenomic variants in 11 genes and were launched on 24 September 2014 for University of Washington inpatient care. Of the 94 participants enrolled in the NEXT Medicine study, 49 had one or more pharmacogenomic variants identified for return.
Conclusion:
Reflections on the process reveal that while incidental findings can be used to generate decision support alerts, substantial resources are required to ensure that each alert is consistent with rapidly evolving pharmacogenomic literature and is customized to fit in the clinical workflow unique to each incidental finding.
Genet Med 17 11, 939–942.
Similar content being viewed by others
Main
Pharmacogenomic data can be used to improve prescribing outcomes and reduce adverse drug events,1 but there are numerous hurdles to implementing it in standard medical care.2 Electronic health records (EHRs) and their associated decision support tools seem to be essential in addressing the challenge of disseminating an individual’s pharmacogenomic profile to his or her providers. A number of institutions have created decision support systems with pharmacogenomic data from targeted gene tests,3,4,5,6,7 but to our knowledge none have used data from exome sequencing.
Exome sequencing is unique compared with targeted genetic tests because it captures a broader range of data that may include incidental findings—“pathogenic or likely pathogenic alterations in genes that are not apparently relevant to a diagnostic indication for which the sequencing test was ordered.”8 A given exome sequencing test may produce thousands of incidental findings with varying degrees of clinical relevance.8,9 Findings with pharmacogenomic implications could be used for patient care, offering the attractive potential of preemptive pharmacogenomics—that is, sequencing data stored in a patient’s EHR and immediately available to help guide care if the patient is ever prescribed an associated medication.
We report a case study of medication alerts generated from pharmacogenomic findings of exome sequencing results using a commercial EHR system.
Materials and Methods
The New EXome Technology in (NEXT) Medicine study is a randomized controlled trial in which patients with a personal and/or family history suspicious for hereditary colon cancer/polyps are randomized to receive usual care or usual care supplemented with exome sequencing. Exome sequencing is performed at the university’s Northwest Clinical Genomics Laboratory. Variants are prioritized for clinical relevance by a clinical molecular geneticist (M.O.D.), a clinical geneticist (G.P.J.), and a genetic counselor (L.M.A.) for presentation to two committees composed of physicians from various specialties, researchers, and ethicists. The nine-member NEXT Medicine Variant Subcommittee reviews challenging variants, determines classifications, and develops reporting language. If there are additional concerns, the Variant Subcommittee consults the 23-member NEXT Medicine Return of Results Committee, which is charged with establishing principles and processes to define an “actionable” gene across the consortium.
Returned pharmacogenomic findings incidental to the primary indication for testing (hereditary colon cancer risk) were required to be clinically actionable with a moderate to strong degree of literature-based evidence. They also were selected by their ability to be captured by current technologies and their relevance for study participants. Selection of incidental findings considered for return of results is outlined in detail elsewhere.10
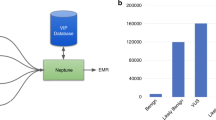
Discrete laboratory reports were created through the University of Washington, Department of Laboratory Medicine to capture pharmacogenomic findings as machine-readable results in the EHR (PowerChart; Cerner, Kansas City, MO). University of Washington Information Technology Services built decision support rules to trigger alerts based on these findings. The rules leveraged preexisting drug–laboratory result rule templates (e.g., penicillin with a penicillin allergy test result). Content for each alert was generated through an iterative process that involved the NEXT Medicine Variant Subcommittee and physicians from multiple specialties. A prototype of the alert was reviewed by physicians, and feedback was incorporated into revisions of the alerts (unpublished data).
An ongoing evaluation plan will allow us to assess the alerts in real medical interactions. Activity will be monitored through automated logging of data regarding when the alert fires, in which department, and how the provider responds to the alert. All providers who encounter the alerts will be asked to complete a survey assessing their perspectives on alert design and content.
Results
The NEXT Medicine study committees established a list of 48 actionable pharmacogenomic variants in 11 genes to return to patients and their providers as incidental findings ( Table 1 ).
In the laboratory result system, incidental findings are structured as paired results: one result is a binary indicator for the presence of abnormal gene activity (e.g., a patient with an actionable CYP2C19 variant is documented as positive for “abnormal CYP2C19 function”); the second result contains text about the variant and its clinical significance. Both results are stored within the lab results section of a patient’s EHR and are machine-readable, allowing them to be utilized by decision support rules engines. In addition, a full report of the returned findings in each participant’s exome is available as two structured, free-text documents that are stored as portable document format files within the EHR.11
The alerts ( Figure 1 ) include a title, variant–drug interaction text, pharmacist contact information, lab result name, and access to Clinical Pharmacogenetics Implementation Consortium guidelines.12 Most alerts fire when a provider submits a medication order for a patient with a relevant gene abnormality documented in the lab result system. The alert allows for three actions: “Cancel Order” eliminates the order and returns to the main order entry screen; “Modify Order” returns to the order entry for the specific drug; and “Override Alert” continues with the existing order. For genes that impact the response to many medications (e.g., HFE), the alert fires when the patient’s record is opened. Alerts are distinct in color and design from drug–drug interaction alerts for added emphasis among health-care providers.
Screenshot of prototype alert for clopidogrel prescription to a patient with a CYP2C19 variant. The prototype alert was built in Cerner Powerchart with a Discern rules engine. It was formatted to appear unique from the drug–drug interaction alerts in the University of Washington inpatient electronic health records system. Concise information about the variant–drug interaction is displayed with a number of recommended actions (e.g., alternate prescription, calling a pharmacist). The guidelines button brings up Clinical Pharmacogenetics Implementation Consortium guidelines in a browser window.
Alerts were launched on 24 September 2014 in the University of Washington Cerner EHR used for inpatient care. Of the 94 participants enrolled in the NEXT Medicine randomized controlled trial, 54 had exome sequencing completed. Of these 54 participants, 49 had one or more pharmacogenomic variants identified for return.
Discussion
Our proof of concept reveals it is possible to use incidental pharmocogenomic findings from exome sequencing to create decision support alerts, which holds exciting prospects for the future of preemptive pharmacogenomic management. During the development process, however, we encountered notable technical and curatorial challenges.
Our binary test result has the benefits of simplicity but fails to capture some information. For example, CYP2C19*2 and CYP2C19*17 are variants with different pharmacokinetic impacts on clopidogrel (slow versus rapid metabolism, respectively) but are documented identically in our laboratory report system. Unless our providers refer to the full lab report, they are alerted only to the fact that the gene is “abnormal.” Documenting lab results with more granularity would allow for more specific alerts, but it would also require a dramatic increase in labor to create and refine the laboratory result ontology.
Laboratory result systems are not the only technology unaccustomed to genomic data; existing EHR systems are similarly unequipped. The constraints of our decision support platform prevented us from including a link to the patient’s full genomic lab report within the alerts—a popularly requested feature among physicians—and from having certain alerts fire only for specialists likely to prescribe the associated medication (e.g., TPMT and thiopurines could be targeted to oncologists). Customizations that can be built into existing decision support tools help reduce unnecessary alerts but demand ample planning and labor costs. Because our warfarin alerts are triggered by four variants from three genes, it was necessary for our information technology staff to generate conditional statements to prevent multiple alerts from firing simultaneously for a patient with more than one such variant. These technical issues are not insurmountable, but they require time and consideration before the alert launch; failure to address them would likely lead to redundant alerts and subsequent alert fatigue.13,14
Evidence curation is another time-consuming task complicated by the rapidly evolving field of pharmacogenomics. Recent trials regarding warfarin exemplify how conflict about the impact of genotyping on patient outcomes and cost-effectiveness exists among various professional societies.15,16,17,18 Thus, we found it prudent to avoid direct commands in alerts. The alert for clopidogrel and CYP2C19 variants states, “Consider using prasugrel or alternative agent,” rather than, “Use prasugrel.”
Our recommendations to others developing decision support for exome sequencing are as follows: anticipate the limits of your institution’s lab result and EHR systems in handling genomic data; account for labor required to customize variant–drug alerts to the clinical context in which they are likely to occur; and, finally, allocate ample resources to gathering, synthesizing, and applying pharmacogenomic evidence on an ongoing basis.
Conclusion
The use of EHRs and their associated decision support tools may be an important way to incorporate the pharmacogenomic incidental findings from exome sequencing into existing clinical workflows. We created a proof of concept of incidental finding–based alerts and explored technical challenges, hurdles in workflow integration, and barriers to content generation. Though technical and labor concerns may currently inhibit the development of exome sequencing decision support systems at many institutions, we anticipate that the advancement of EHR systems and the solidification of centralized, pharmacogenomic knowledge bases will make this a promising technology in the near future.
Disclosure
The authors declare no conflict of interest.
References
Green ED, Guyer MS ; National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204–213.
Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther 2012;92:467–475.
Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014;89:25–33.
Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 2014;21(e1):e93–e99.
Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014;166C:45–55.
O’Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care—initial results of the University of Chicago “1,200 Patients Project.” Am J Med Genet C Semin Med Genet; 2014;166C:68–75.
Shuldiner AR, Palmer K, Pakyz RE, et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet 2014;166C:76–84.
Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574.
Burke W, Antommaria AH, Bennett R, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med 2013;15:854–859.
Berg JS, Amendola LM, Eng C, et al.; Members of the CSER Actionability and Return of Results Working Group. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med 2013;15:860–867.
Dorschner MO, Amendola LM, Shirts BH, et al. Refining the structure and content of clinical genomic reports. Am J Med Genet C Semin Med Genet 2014;166C:85–92.
Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011;89:464–467.
Paterno MD, Maviglia SM, Gorman PN, et al. Tiering drug-drug interaction alerts by severity increases compliance rates. J Am Med Inform Assoc 2005;16:40–46.
Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform 2012;45:1202–1216.
Pirmohamed M, Burnside G, Eriksson N, et al.; EU-PACT Group. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013;369:2294–2303.
Kimmel SE, French B, Kasner SE, et al.; COAG Investigators. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013;369:2283–2293.
Verhoef TI, Ragia G, de Boer A, et al.; EU-PACT Group. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med 2013;369:2304–2312.
Zineh I, Pacanowski M, Woodcock J. Pharmacogenetics and coumarin dosing–recalibrating expectations. N Engl J Med 2013;369:2273–2275.
Acknowledgements
The authors acknowledge the assistance of Aidan Garver-Hume (University of Washington Information Technology Services), Tony Shaver (University of Washington School of Pharmacy), and Chuck Rohrer (University of Washington Laboratory Medicine). This study was supported by National Institutes of Health, National Human Genome Research Institute, and National Cancer Institute grants U01HG006507, U01HG006375, and U01HG007307; National Institute of Translational Health Sciences grant UL1TR000423; and National Institutes of Health and National Center for Advancing Translational Sciences grant TL1 TR000422.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishimura, A., Shirts, B., Dorschner, M. et al. Development of clinical decision support alerts for pharmacogenomic incidental findings from exome sequencing. Genet Med 17, 939–942 (2015). https://doi.org/10.1038/gim.2015.5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.5
Keywords
This article is cited by
-
Integrating clinical decision support systems for pharmacogenomic testing into clinical routine - a scoping review of designs of user-system interactions in recent system development
BMC Medical Informatics and Decision Making (2017)
-
Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences
npj Genomic Medicine (2016)
-
Returning pharmacogenetic secondary findings from genome sequencing: let’s not put the cart before the horse
Genetics in Medicine (2015)