Abstract
Purpose:
Genetic testing has shifted from academic laboratories with expertise in specific genes to commercial laboratories that offer tests of a diverse array of genes. The purpose of this comparative study was to determine whether one academic laboratory’s model of variant interpretation is similar to that of several commercial laboratories.
Methods:
The Collagen Diagnostic Laboratory (CDL) received, over a 14-month period, 38 requests to interpret variants originally identified by an outside laboratory (OL). The interpretations by the OL and CDL were compared and discrepancies were assessed.
Results:
Interpretations from the OL and CDL were concordant in 11 inquiries (29%); discrepancies were moderate in 11 instances (29%) and significant in 16 (42%). Factors that caused discrepancies included the following: (i) private data were not shared in a public database (n = 9); (ii) publicly available allele frequency data were not referenced and used as evidence (n = 5); and (iii) important aspects of protein structure and function were not taken into account (n = 13).
Conclusion:
Comprehensive interpretation of sequence variants depends on good functional tests and well-curated variant databases. Provision of clinical information to the clinical laboratory, mandatory submission of identified variants with phenotype data to common resources, and collaboration between clinical laboratories and recognized experts is likely to improve consistency in variant interpretation among clinical laboratories.
Genet Med 18 1, 20–24.
Similar content being viewed by others
Introduction
Identification of a gene sequence variant of unknown significance (VUS) during clinical testing, even in well-studied genes, is commonplace.1,2 Laboratory professional organizations involved in clinical genetic testing offer recommendations to standardize the interpretation and reporting of sequence variants by creating classification categories and standards,3,4 though implementation of these guidelines is not mandatory. Regardless of the method with which the variant is identified, interpretation often combines an assessment of variant frequency in presumably unaffected populations (e.g., the Exome Aggregation Consortium,5 the Exome Variant Server,6 the 1000 Genomes Project7); prior reports of pathogenicity; conservation across species; familial segregation; in silico analysis of predicted impact (e.g., Sorting Intolerant From Tolerant (SIFT),8 Poly-Phen2 (ref. 9)), and clinical judgment. Ideally, variant interpreters at different clinical genetics laboratories will come to the same assessment of clinical significance if they use the same interpretative tools, data sources, and variant-classification guidelines and approaches. Nonetheless, variability in the interpretation of genetic alterations can occur between different clinical laboratories.10 Data sharing between laboratories could help minimize discrepant variant interpretations, such as through publicly available databases like ClinVar11; at present, however, the degree of laboratory participation seems to be inadequate, and the quality of the submitted variant interpretations is variable.
As a university-based clinical and research laboratory specializing in heritable connective-tissue disorders, the Collagen Diagnostic Laboratory (CDL) at the University of Washington routinely receives inquiries from genetic counselors and clinical geneticists regarding a variant identified in an outside laboratory (OL). These requests typically involve situations in which the patient had been tested during evaluation for a suspected heritable connective-tissue disorder and a variant was identified in a gene that was among those studied in the CDL. In some cases, variants were identified through whole-exome sequencing. In each instance the clinician sought additional information or reinterpretation from CDL as a “second opinion” on the variant to provide genetic counseling and recommendations to the patient and family. During the course of providing this routine clinical service, it became apparent that in some cases there were discrepancies between variant interpretations issued by the OL and evaluation by CDL. To better understand factors underlying differing interpretations, we set out to study and characterize our experience with interpretation of variants originally identified at an OL and submitted by clinicians to CDL for inquiry.
Materials and Methods
Sample ascertainment
Between January 2013 and March 2014, 38 inquiries of variant interpretation from clinicians were received by e-mail or phone by either a laboratory genetic counselor or a laboratory director. The variant description was recorded and a copy of the de-identified OL report was requested to confirm the variant description and original interpretation. Demographic data were recorded at the disposition of the inquiry (Supplementary Table S1 online).
CDL investigation of variants
Upon submission of a request for interpretation, the variant was investigated by a credentialed laboratory genetic counselor and laboratory director. If a variant was previously identified in the CDL, the most recent interpretation was offered after confirmation that there were no updated informational resources. If the variant had not been previously identified in the CDL, the variant was investigated per a protocol that included searching publicly available loci-specific and population databases, referencing conservation and assessing predicted protein effects (Supplementary Chart S1 online). The referenced databases included the Osteogenesis Imperfecta/Ehlers-Danlos Syndrome International Database,12,13 the Universal Mutation Database FBN1 mutations database,14 the Universal Mutation Database TGFBR1 and TGFBR2 databases,15 and the National Center for Biotechnology Information ClinVar database.11 At the conclusion of the investigation, variants were classified as one of the following categories based on clinical assessment of the available evidence: pathogenic, likely pathogenic, VUS, likely benign, or benign.
Comparison of interpretation and identification of discrepancies
To compare the interpretation of variants between the CDL and the OL, we reviewed the report issued by the OL and provided by the clinician to identify factors included in the variant interpretation. In some instances only the result (i.e., not the full interpretation) was available for analysis. The degree of interpretative discrepancies was categorized as follows:
-
1. Significant—the classification category of the variant assigned by the OL and CDL differed by two or more steps in clinical significance (e.g., benign versus VUS or pathogenic versus VUS).
-
2. Moderate—the classification category of the variant assigned by the OL and CDL differed by one step in clinical significance (e.g., pathogenic versus likely pathogenic or likely pathogenic versus VUS).
-
3. None—the classification category of the variant assigned by the OL and CDL was congruent (e.g., VUS versus VUS).
If interpretations differed between the OL and CDL, factors that could contribute to the discrepancy were identified and categorized as one of the following:
-
1. Private data—variant and/or additional evidence of clinical significance was not available to the OL because the CDL had not submitted variant and/or additional investigative studies to publicly accessible genetic databases.
-
2. Public data not referenced—variant and/or additional information that could aid in interpretation was available in a publicly accessible genetic database(s) but was not explicitly referenced in the OL’s interpretation.
-
3. Predicted protein consequence not referenced—predicted biologic significance (i.e., the effect of the variant on protein structure and function) was not referenced in the OL’s interpretation.
Results
Summary of inquiries
Inquiries were received from genetic counselors (n = 25), clinical geneticists (n = 12), and an orthopedist (n = 1). The original genetic testing was completed in five private commercial laboratories (20 sample inquiries) and six academic laboratories (12 sample inquiries); six were from unidentified laboratories. The genes of inquiry were COL3A1 (n = 21), COL1A1 (n = 7), COL1A2 (n = 5), FBN1 (n = 2), TGFBR1 (n = 2), and TGFBR2 (1). The types of variants included missense (n = 28), intronic (n = 6), in-frame duplications (n = 2), and synonymous (n = 1). The methodology through which the variant was identified by the OL was exome sequencing (n = 6), next-generation sequencing panel (n = 17), single-gene Sanger sequencing (n = 8), or not provided (n = 7). The amount of time that elapsed between the OL’s interpretation and the inquiry to the CDL was a median of 11 days (range 1–60 days) (n = 12).
Comparison of interpretation of the identified variant between the OL and CDL
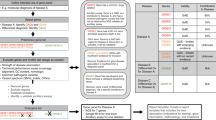
The OL and CDL had concordant variant interpretations in 11 cases (29%) ( Figure 1 ). A significant discrepancy in interpretation occurred in 16 cases (42%). Significant discrepancies were instances in which the difference in interpretation was two or more classification categories and affected the clinical genetic diagnosis. Moderate discrepancies occurred in 11 cases (29%). These included instances in which the difference in interpretation was a single classification category and resulted in a moderate impact to clinical care in the form of additional genetic testing, delay in diagnosis, or the need for segregation studies.
Comparison of Collagen Diagnostic Laboratory (CDL) and Outside Laboratory (OL) variant interpretations (a) Classification of the predicted clinical impact resulting from discrepancy in interpretation. (b) Comparison of the number of interpretations, by interpretation categories, in the CDL (gray) and the OLs (dark gray).
Evaluation of the discrepancy
To understand the reason for discordance between the OL and CDL interpretations, the OL reports were reviewed to determine the sources and resources used. Based on data provided in 24 reports available for analysis, of which 16 were for variants with discrepant interpretations, the predicted protein consequence of the variant was not referenced in the OL’s interpretation in eight cases, publicly available data were not referenced in the OL’s interpretation in two cases, and private data accessible only to the CDL explained the discrepancy in six cases. In 14 remaining cases, 11 of which were variants with discrepant interpretations, the OL report was not available but the OL’s interpretation was provided to us by the clinician. Among these, the effect of the variant on protein structure or function did not seem to be referenced in five cases, data accessible only to CDL could have changed the interpretation in three cases, and there was no apparent reference to publicly available databases in three cases. Overall, of the 27 discrepant results, 33% seemed to be influenced by a lack of access by the OL to private CDL data, 19% seemed to be influenced by a lack of reference to publicly available data, and 48% seemed to be influenced by a lack of reference to the current understanding of the biology of the investigated gene. (see Supplementary Table S2 online)
Discussion
For most of the variants in this study, the CDL was consulted because the clinician was uncertain about the clinical implication of the interpretation by the OL. In this small set of 38 instances, discrepancies in the interpretation of variants between OLs and the CDL occurred in more than half the samples. Of most concern is that nearly 50% of the discrepant interpretations are predicted to significantly affect the molecular diagnosis or clinical care recommendations. Although we do not know whether our experience with this limited set of genes and examples is generalizable, it raises concern and warrants further investigation. Uncertainty in laboratory results increases the burden of interpretation for the clinician and may lead to misdirection in medical care for the patient, at significant cost.16 In just over half (13 of 25) of cases classified by an OL as a VUS, the CDL interpreted the variant as either definitively benign or pathogenic. In one case a variant interpreted as pathogenic by an OL was classified as benign by the CDL; in 13 other cases either moderate or significance discrepancies in classification occurred. This difference in variant interpretation makes it important to understand why different assessments were reached and to identify means by which more consistent interpretation across laboratories can be attained.
One factor that seemed to influence discrepant variant interpretation was that data accessible to the CDL from prior testing and investigation were not in public genetic databases. At present, laboratories are encouraged to submit the de-identified variants and clinical correlates to publicly available databases to allow for universal access by testing laboratories and clinicians. Four of the 12 OLs (30%) that performed the original variant interpretations investigated in this study have submitted some gene sequence variants into ClinVar, as has the CDL. Of the genes interrogated in the OL inquiries, 80% of recorded ClinVar pathogenic entries were submitted by the CDL, which reflects a low submission rate for genes associated with heritable connective-tissue disorders to date by other laboratories.
With time, the increased availability of variant data should decrease discrepant interpretations among laboratories that result because data are available to only one source. However, submission of all accrued variants from prior years and decades of testing faces the logistical challenge of insufficient laboratory resources to allocate to the task of manual curation and submission. In smaller academic laboratories especially, dedicating uncompensated personnel time to submit archives of variant data to public databases comes at a significant cost. At the CDL, this effort is undertaken piecemeal over time, which is one factor that contributed to discrepant interpretations of variants identified by OLs.
About a quarter of discrepant interpretations seemed to result from a lack of reference to data that are in publicly available databases. We could not tell whether the databases were not used, their significance was not appreciated, or the activity simply was not reported because there is not a standard description among laboratories of the resources and references used. Annotation of tools and databases used in variant interpretation should be provided and may help clinicians seeking second interpretations and assist other laboratories when testing relatives.17 If locus-specific variant databases can be assembled in a standardized fashion (see, for example, the Leiden Open Variation Database format in the Mutation Database of Osteogenesis Imperfecta and Ehlers Danlos syndrome13) and interfaced with centralized databases for efficient access and download of all variants, the ability to use them in an automated fashion may result. This should improve the concordance among laboratory interpretations as the shared information on which variant interpretation is based increases.
The challenge of accurate interpretation of variants identified on a large scale by exome and genome sequencing is a focus of clinical laboratory directors and policymakers.18 Current American College of Medical Genetics and Genomics guidelines (and the revision of those guidelines that is under way) allow for classification of pathogenicity if the variant is of the type expected to result in disease, even if novel. However, especially for missense variants, the ability to assess predicted protein consequence requires familiarity with protein structure and critical functional domains. In the absence of an intimate understanding of protein biology and knowledge of common mutational mechanisms in the associated gene, the significance of novel missense variants can be difficult to assess. For example, the triple helical domain of fibrillar collagens (types I, II, III, V, and XI) contain an uninterrupted string of Gly-X-Y triplets that extends for more than 1,000 residues. To date, virtually all substitutions for the canonical glycine residues result in gene-specific phenotypes. Five of the 38 unique variants submitted for inquiry in this study were substitutions for canonical glycine that are expected to result in disease based on fibrillar collagen biology and the common mutation mechanism in these genes. Nonetheless, four were interpreted as variants of uncertain significance presumably because they were novel or not publicly reported with sufficient supporting evidence to conclude pathogenicity. Because between 60% and 80% of disease-causing mutations in the three collagen genes with which we have experience (COL1A1, COL1A2, and COL3A1) are family-specific, the absence of a report in the literature or a database is an expected finding. Classification of a substitution for glycine within the triple helical domain as a VUS shifts the burden to interpret the variant to the clinician, delays diagnosis, and may lead to expensive additional testing. Although in silico analysis may be a helpful tool to guide and improve variant interpretation, more specific communication of information used in these programs, such as conservation and the nature of the substituting residue, can be informative because the programs are not vetted for clinical use and have recognized short-comings.19,20 The next generation of such tools, like Structural Disruption Score (SDS),21 that predict the protein consequence of a particular nucleotide variation may provide more generalizable outcomes but do need to be validated against existing variant and clinical databases. When our understanding of the molecular blueprint of unique gene families is translated into a computational algorithm that includes the extant knowledge from large, accurate genotype–phenotype databases, the proportion of true VUSs will diminish.
As diagnostic laboratory test menus continue to expand in response to clinical demand, and with increased uptake of whole-exome/-genome sequencing, every laboratory director being familiar with the structure and function of each gene and protein in which variants are identified is untenable. Collaboration between diagnostic laboratories and disease-specific experts or consortiums may improve the ability to interpret the significance of variants and reduce the rate of reporting VUSs, such as has been undertaken by the Evidence-based Network for the Interpretation of Germline Mutant Alleles.22 In addition, participation in more generalized organizing bodies, such as the International Collaboration for Clinical Genomics,23 may also aid in this endeavor.
Limitations to our findings and the generalizability of our conclusions include the small number of inquiries, the absence of physical laboratory reports and the lack of details about OL interpretations in several instances, and the small family of heritable connective-tissue disorder genes interrogated. Because we were approached by clinicians who received the results from an OL, we were not privy to the full extent of information taken into consideration by the OL in its interpretation of the variant (some of which may not have been included in the reported interpretation). For this reason, we cannot exclude the possibility that other factors contributed to the discrepancy of interpretations in some instances, such as private data available only to the OL. While these limitations are important to consider in the generalization of our findings to variant interpretation on a genomic scale, this study provides insight into some factors that influence discrepant interpretations of variants by different clinical laboratories, as well as possible steps toward reducing those discrepancies.
In conclusion, our study suggests that discrepancies in variant interpretation occur between accredited clinical laboratories and that standardization of variant interpretation will be aided by expansion of the catalog of variants that are publicly available, standards for the use of investigative algorithms, and inclusion of predicted protein consequences. We propose the following measures to improve report accuracy and consistency in variant interpretation in clinical genetic testing.
-
1. Standardize the report format to include all the evidence considered in the variant interpretation to promote transparency of the investigative algorithm used to determine clinical significance.
-
2. Modify the current recommendations to allow the use of predictive algorithms, when validated, to provide better assessment of the effects of variants.
-
3. Mandate that clinical laboratories contribute all variants and clinical correlates to publicly accessible genetic databases to improve availability of data and include this obligation as part of CLIA/College of American Pathologists certification quality assurance measures to provide an incentive for laboratory compliance.
-
4. Create an avenue for collaboration between clinical laboratories and disease- and gene-specific experts to aid in the interpretation and curation of variants identified in genes that they know well.
-
5. Create and support consortiums or networks that can define gene-specific rules for variant interpretation and support expert curation of variants for clinical use.
Disclosure
The authors declare no conflict of interest.
References
Calò V, Bruno L, Paglia LL, et al. The Clinical Significance of Unknown Sequence Variants in BRCA Genes. Cancers (Basel) 2010;2:1644–1660.
Shirts BH, Jacobson A, Jarvik GP, Browning BL. Large numbers of individuals are required to classify and define risk for rare variants in known cancer risk genes. Genet Med 2014;16:529–534.
Richards CS, Bale S, Bellissimo DB, et al.; Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 2008;10:294–300.
Claustres M, Kožich V, Dequeker E, et al.; European Society of Human Genetics. Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic). Eur J Hum Genet 2014;22:160–170.
Exome Aggregation Consortium (ExAC), Cambridge, MA, 2015. http://exac.broadinstitute.org/. Accessed 15 January, 2015.
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, 2014. http://evs.gs.washington.edu/EVS/. Accessed 15 January, 2015.
1000 Genomes Project Consortium; Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012;7:e46688.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20.
Tsuchiya KD, Shaffer LG, Aradhya S, et al. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet Med 2009;11:866–873.
Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42(Database issue):D980–D985.
Dalgleish R. A Database of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome Variants, 1997. http://www.le.ac.uk/ge/collagen/. Accessed 15 January, 2015.
Dalgleish R. The Human Collagen Mutation Database 1998. Nucleic Acids Res 1998;26:253–255.
Collod-Béroud G, Le Bourdelles S, Ades L, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat 2003;22:199–208.
Frederic MY, Hamroun D, Faivre L, et al. A new locus-specific database (LSDB) for mutations in the TGFBR2 gene: UMD-TGFBR2. Hum Mutat 2008;29:33–38.
Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA 2014;311:1035–1045.
Dorschner MO, Amendola LM, Shirts BH, et al. Refining the structure and content of clinical genomic reports. Am J Med Genet C Semin Med Genet 2014;166C:85–92.
Cutting GR. Annotating DNA variants is the next major goal for human genetics. Am J Hum Genet 2014;94:5–10.
Tavtigian SV. Comparison of programs for in silico assessment of missense substitutions. Hum Mutat 2011;32:v.
Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat 2011;32:661–668.
Preeprem T, Gibson G. SDS, a structural disruption score for assessment of missense variant deleteriousness. Front Genet 2014;5:82.
Spurdle AB, Healey S, Devereau A, et al.; ENIGMA. ENIGMA–evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 2012;33:2–7.
International Collaboration for Clinical Genomics, 2014. http://iccg.org/. Accessed 15 January, 2015.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
(ZIP 44 kb)
Rights and permissions
About this article
Cite this article
Pepin, M., Murray, M., Bailey, S. et al. The challenge of comprehensive and consistent sequence variant interpretation between clinical laboratories. Genet Med 18, 20–24 (2016). https://doi.org/10.1038/gim.2015.31
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.31
Keywords
This article is cited by
-
From the laboratory to the clinic: sharing BRCA VUS reclassification tools with practicing genetics professionals
Journal of Community Genetics (2018)
-
A variant by any name: quantifying annotation discordance across tools and clinical databases
Genome Medicine (2017)
-
Integration of Clinical Genetic Testing in Cardiovascular Care
Current Genetic Medicine Reports (2016)