Abstract
Purpose:
We aimed to determine the prevalence and phenotypic spectrum of NOTCH1 mutations in left-sided congenital heart disease (LS-CHD). LS-CHD includes aortic valve stenosis, a bicuspid aortic valve, coarctation of the aorta, and hypoplastic left heart syndrome.
Methods:
NOTCH1 was screened for mutations in 428 nonsyndromic probands with LS-CHD, and family histories were obtained for all. When a mutation was detected, relatives were also tested.
Results:
In 148/428 patients (35%), LS-CHD was familial. Fourteen mutations (3%; 5 RNA splicing mutations, 8 truncating mutations, 1 whole-gene deletion) were detected, 11 in familial disease (11/148 (7%)) and 3 in sporadic disease (3/280 (1%)). Forty-nine additional mutation carriers were identified among the 14 families, of whom 12 (25%) were asymptomatic. Most of these mutation carriers had LS-CHD, but 9 (18%) had right-sided congenital heart disease (RS-CHD) or conotruncal heart disease (CTD). Thoracic aortic aneurysms (TAAs) occurred in 6 mutation carriers (probands included 6/63 (10%)).
Conclusion:
Pathogenic mutations in NOTCH1 were identified in 7% of familial LS-CHD and in 1% of sporadic LS-CHD. The penetrance is high; a cardiovascular malformation was found in 75% of NOTCH1 mutation carriers. The phenotypic spectrum includes LS-CHD, RS-CHD, CTD, and TAA. Testing NOTCH1 for an early diagnosis in LS-CHD/RS-CHD/CTD/TAA is warranted.
Genet Med 18 9, 914–923.
Similar content being viewed by others
Introduction
Left-sided congenital heart disease (LS-CHD) represents a group of highly heritable congenital heart defects, including bicuspid aortic valve (BAV), aortic valve stenosis (AVS), coarctation of the aorta (COA), and hypoplastic left heart syndrome (HLHS). LS-CHD includes the previously used term “left ventricular outflow tract obstruction.”1,2,3,4,5 BAV may be asymptomatic and undetected in infancy, but it has a risk for serious complications and sudden cardiac death later in life.6,7 HLHS is a severe disease that requires extensive surgical corrections and may lead to death at a young age.8,9 LS-CHD often presents as a nonsyndromic condition but has been reported in more than 200 syndromes; in many of these the associated gene or chromosomal region is known, for example, Jacobsen syndrome (11q23 deletion), Turner syndrome (monosomy X), Kabuki make-up syndrome (MLL2, KDM6A), and Rieger syndrome (PITX2, FOXC1).10,11,12,13 In nonsyndromic LS-CHD, only a few genes are known to be associated with the disease (NOTCH1, GJA1, NKX2-5, GATA5, SMAD6, MYH6), and the number of patients with mutations in these genes is small.14,15,16,17,18,19,20,21 NOTCH1 is the only gene reported with truncating mutations segregating with LS-CHD, most often BAV, and/or early-onset calcific AVS.
The NOTCH1 gene codes for the transmembrane receptor protein NOTCH1, which is part of the NOTCH signaling pathway. NOTCH signaling is evolutionary conserved and plays an important role in embryonic development by influencing cell fate.22 NOTCH signaling mediates short-range intercellular communication: the transmembrane receptors NOTCH1–4 interact with ligands (Delta-like 1, 3, and 4, and Jagged (JAG) 1 and 2) from neighboring cells. After ligand binding, the receptor is cleaved and an intracellular domain enters the nucleus, where it interacts with DNA-binding proteins.23 Downstream targets of NOTCH1 are among the Hes (hairy enhancer of split) and Hrt (Hes-related) families of genes.24,25
Activation of NOTCH1 represses differentiation of embryonic stem cells into cardiomyocytes and stimulates the endocardial epithelial-to-mesenchymal transition, which is important in the process of cardiac valve formation.26,27 In addition, NOTCH1 haploinsufficiency leads to calcification of the aortic valve by dysregulating the downstream transcription of genes involved in osteogenesis, inflammation, and oxidative stress.28,29 Germ-line truncating mutations in NOTCH1 were first reported as segregating in two families with mainly aortic valve disease and two patients with conotruncal heart disease (CTD) (Fallot’s tetralogy and double-outlet right ventricle).14 One of these mutations was also reported in a patient with a stenotic tricuspid aortic valve.30 In five series of patients with LS-CHD screened for NOTCH1 mutations, only two new truncating mutations, one RNA-splicing mutation, and several possibly pathogenic missense variants were reported.15,31,32,33,34 Because these series are small and the number of mutations reported is limited, the role of NOTCH1 in LS-CHD is still unclear.
In this article the phenotypic spectrum and pedigrees of patients with clearly pathogenic NOTCH1 mutations are presented, as well as data on nonsynonymous variants. This information will help clinicians to make decisions upon DNA testing and when counseling patients on risk profiles for their relatives and their offspring.
Materials and Methods
Patients
Patients with LS-CHD referred for genetic counseling to one of three participating university hospitals in the Netherlands between 1 January 2006 and 1 January 2014 were included in the study. This is a selected population, which can be expected to have a higher prevalence of familial disease than described earlier in LS-CHD (20–35%, depending on the definition of familial disease).4 Intrauterine deaths and terminations of pregnancy were also included. All patients had a detailed cardiac evaluation by a (pediatric) cardiologist, including electrocardiographym and echocardiography/Doppler imaging. Magnetic resonance imaging was performed if the aortic arch could not be visualized by echocardiography. Cardiac diagnoses included were BAV, AVS, aortic valve insufficiency, COA (with or without BAV), HLHS, or other left-sided cardiac diseases, including subvalvular or supravalvular aortic stenosis, hypoplastic aortic arch, interruption of the aorta (type B), and mitral valve anomalies. In patients with combined lesions, the primary diagnosis was defined as the most relevant anomaly, so, if a BAV and a COA were present, the diagnosis was COA. All normally functioning, stenotic, or insufficient BAVs were labeled BAV. HLHS was defined as underdevelopment of the left ventricle and ascending aorta, together with anomalies of the mitral and/or aortic valve. A complete physical examination was performed, and a detailed family history was taken by a clinical geneticist. Patients with major extracardiac malformations or known syndromes were excluded. Familial LS-CHD was defined as LS-CHD in the proband and LS-CHD or any congenital heart disease (CHD) in a first-degree relative, or LS-CHD in the proband and LS-CHD or right-sided CHD (RS-CHD)/CTD in a second- or third-degree relative. The RS-CHD/CTD group included pulmonary valve stenosis, pulmonary atresia, Fallot’s tetralogy, and truncus arteriosus. Echocardiography was offered to first-degree relatives as described previously.4
Sequence analysis of the NOTCH1 gene
Mutation analysis of the coding exons and flanking intronic sequences of the NOTCH1 gene (NM_0176173) was carried out using flanking intronic primers (primer sequences are available upon request). Polymerase chain reaction (PCR) was performed in a total volume of 15 µl containing 10 µl AmpliTag Gold Fast PCR Master Mix (Applied Biosystems, Bleiswijk, the Netherlands), 1.5 pmol/µl of each primer (Eurogentec, Seraing, Belgium), and 2 µl (40 ng/µl) genomic DNA. To the PCR mix of exon 1 was added 10% dimethyl sulfoxide. The samples were PCR amplified on GeneAmp 9700 (Applied Biosystems, Bleiswijk, the Netherlands) (see Supplementary Methods online for conditions). The PCR products were purified with ExoSAP-IT (Amersham Pharmacia Biotech, Piscataway, NJ) and subjected to direct sequencing on a Prism 3730XL DNA analyzer (Applied Biosystems, Bleiswijk, the Netherlands) using the specific primers.
Array-comparative genomic hybridization analysis
To detect deletions, array-comparative genomic hybridization analysis was performed using an 180K oligo array (custom design ID 23363; Agilent Technologies, Santa Clara, CA). A mix of 40 healthy male or 40 female DNA samples was used as a reference (sex-matched). Procedures were performed according to the manufacturer’s protocol. Data were extracted using Feature Extraction software version 9.1. This analysis was not included in the genetic workup by all clinical geneticists. Data were available for 180 patients, and these were analyzed for deletions in the chromosome 9q34 region, which contains the NOTCH1 gene.
Classification of mutations
Mutations that cause premature truncation (nonsense and frameshift mutations) or a complete deletion of the protein were classified as pathogenic. Mutations within two base pairs upstream or downstream from the exon were presumed to affect RNA splicing and therefore also considered pathogenic. Alamut (version 2.3) missense prediction and splicing prediction modules from Interactive Biosoftware (http://www.interactive-biosoftware.com) were used to predict pathogenicity. The missense prediction module includes Align GVGD, SIFT, PolyPhen-2, and MutationTaster; the splicing module includes SpliceSiteFinder, MaxEntScan, NNSplice, GeneSplicer, and Human Splicing Finder. Variants with a minor allele frequency (MAF) >0.01 were considered polymorphisms and were not registered; (allele frequency data were derived from the National Heart, Lung, and Blood Institute Exome Sequencing Project for European Americans (http://evs.gs.washington.edu/EVS/) and from the Exome Aggregation Consortium browser for Europeans (non-Finnish) (http://exac.broadinstitute.org)).
See the legend of Supplementary Table S1 online for the rules used to classify the nonsynonymous variants.
Complementary DNA analysis of c.3787C>T;p.Arg1263Cys and c.1670-7G>A, potential RNA splicing mutations
All missense variants, synonymous DNA variants, and intronic variants farther than two base pairs upstream or downstream from the exon were analyzed in silico for their effect on RNA splicing. Two variants—c.3787C>T;p.Arg1263Cys and c.1670-7G>A—were predicted to introduce a new splice donor site and were therefore tested. Total RNA was extracted from peripheral blood using the RNABee procedure (Cinna Biotecx, Friendswood, TX), and complementary DNA (cDNA) was obtained using the GoScript Reverse Transcription System (Promega, Leiden, the Netherlands).
Primers amplifying a product from the exon 21–exon 22 transition site to exon 24 TGCAAGTGCGTGGCCGGCTACCA (forward) and CCATTCTTGCAGGGCTTGCCTTT (reverse) were used to characterize the cDNA sequence around the c.3787 C>T mutation in exon 23.
Primers amplifying a product from the exon 9–exon 10 transition site to exon 12, GGGCTTCACTGGGCATCTG (forward) and GGCACACTCGTAGCCATCG (reverse) were used to characterize the cDNA sequence around the mutation in intron 10, c.1670-7G>A.
The PCR products were loaded on to a 3% agarose gel, purified with ExoSAP-IT, and subjected to direct sequencing to confirm the presence of an aberrant transcript.
Statistical analysis
We used SPSS for Windows (version 20; SPSS, Inc., Chicago, IL) for the statistical analyses. A P value <0.05 was considered statistically significant. Interquartile range was used to show statistical dispersion.
Results
Patients
A cohort of 428 probands (286 male, 142 female) with nonsyndromic LS-CHD, including 31 intrauterine deaths/terminated pregnancies, was included. The median age (terminated pregnancies excluded) was 10 years (interquartile range, 2–30 years). In 148 of 428 probands (35%) the heart defect was familial. Cardiac phenotypes are described in Table 1 .
Truncating and RNA-splicing mutations and whole-gene deletions
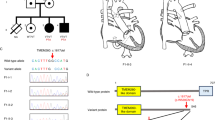
In total, we detected 14 novel clearly pathogenic mutations among 428 probands (3%): 8 truncating (nonsense or frameshift) mutations, 5 RNA-splicing mutations, and 1 whole-gene deletion. ( Figure 1 and Table 2 ) Two variants (c.3787 C>T, p.Arg1263Cys and c.1670-7G>A) were predicted to introduce a new splice donor site and were therefore tested via cDNA analysis. c.3787 C>T, p.Arg1263Cys: cDNA sequencing confirmed that a new splice donor site was introduced in exon 23, causing a deletion of 120 base pairs at the 3′ site of the exon, resulting in p.Glu1262_Gly1301del.
NOTCH1 protein, position of truncating and RNA-splicing mutations. ↓Truncating and RNA-splicing mutations. ANK, ankyrin repeats; EGF, epidermal growth factor; HD, heterodimerization domain; LNR, Lin/Notch repeats; PEST, PEST domain; RAM, RBPjk-association module; TAD, transcriptional activation domain; TM, transmembrane domain.
c.1670-7G>A: cDNA sequencing confirmed that a new splice acceptor site was introduced five nucleotides downstream from the original splice acceptor site (r.1670-5_1670-1ins), causing a frameshift p.Gly557fs. The 14 clearly pathogenic mutations were found in probands with BAV (n = 7, families A, D, G, I, J, M, and N, as shown in Table 3 ); AVS (n = 3, families B, C, and F); COA with BAV (n = 2, families E and H); and HLHS (n = 2, families K and L). Eleven mutations were detected in 148 probands with familial LS-CHD (7%); six were truncating and five were RNA-splicing mutations. Three mutations were detected in 280 nonfamilial cases (1%) (families I, K, and L): two were truncating mutations (one de novo in a patient with HLHS and one inherited from an unaffected parent in a patient with BAV), and one was a de novo whole-gene deletion in a patient with HLHS (detected by array-comparative genomic hybridization).
Phenotypes in families with clearly pathogenic mutations
Relatives of the 14 probands who had pathogenic mutations were subsequently tested; in these families we identified 49 new mutation carriers, 8 of whom were obligate carriers. Thirteen other relatives had a congenital heart defect, but DNA was not available for testing.
Pedigrees are presented in Figure 2 and the phenotypes are summarized in Table 3 . BAV was present in 19/63 mutation carriers (probands included) (30%), AVS without BAV was present in 10/63 mutation carriers (16%), aortic valve insufficiency without BAV in 5/63 (8%), COA in 2/63 (3%), and HLHS in 2/63 (3%). Nine mutation carriers (14%) presented with an RS-CHD/CTD; in three of those this occurred together with an LS-CHD. Sudden death without a previous cardiac diagnosis (at ages 37, 39, and 53 years) was reported in two obligate mutation carriers and one twin sister of a mutation carrier. One deceased obligate carrier was reported by the family to have a “valve insufficiency,” but medical records were not available. Six mutation carriers had thoracic aortic aneurysm (TAA) larger than 40 mm; this was associated with BAV in four and with AVS without documented BAV in two. Seven of the 13 affected relatives who were not tested had AVS; 2 of these had documented BAV (1 with associated TAA), and 1 patient with AVS had associated total anomalous pulmonary venous return. Mitral valve disease was present in three untested relatives, pulmonary valve atresia with Fallot’s tetralogy or ventricular septal defect (VSD) in two, VSD in one, and VSD and atrial septal defect II in one. Twelve of the 49 mutation carriers (25%) were asymptomatic (confirmed by echocardiography in all), so apparently the mutation was nonpenetrant in these individuals. The age range of these relatives was 16–75 years; the younger ones might still develop calcification of the aortic valve and/or TAA in the future. Nonpenetrance occurred in four patients with maternally inherited NOTCH1 mutations and in four patients with paternally inherited mutations, and no DNA was available from the parents of four mutation carriers with a normal echocardiogram (family B patient I.2, family I patient II.1, family J patient I.1, and family N patient II.5).
Pedigrees of patients with truncating and RNA-splicing mutations in NOTCH1 f rom 14 families of probands with mutations. The mutations in families K and L were de novo. See Table 3 for a description of the phenotypes.
In patient II.1 from family H, sustained ventricular tachycardia was registered in the hospital and treated with ablation. In her daughter (patient III.1), who complained of palpitations, no arrhythmia was registered on Holter registration.
Nonsynonymous variants
Apart from these pathogenic mutations, 24 nonsynonymous variants (11 novel) were detected in 35 of the 428 patients (8%). These variants are listed in Supplementary Table S1 online, with data about the phenotype, outcome of the prediction programs, number of controls tested, and segregation of the variants in the families. In one patient with BAV and TAA, two nonsynonymous variants were present. The parents were available for testing in 22 patients. We found 18 carrier parents were not affected, and this was confirmed with echocardiography in 16 of them. In the family with variant p.(Cys344Ser), however, the unaffected carrier mother had two close relatives who were reported by the family to have had a congenital heart defect but were deceased (no medical files or DNA were available). In two families the variant was not present in an affected relative, suggesting that these variants (p.(Lys1461Arg) and p.(Met1669Arg)) are not the main disease-causing factors. Based on the segregation observed, prediction programs, and data from controls, we consider 12 variants to be probably benign and 2 variants to be probably pathogenic (p.(Cys344Ser) and p.(Asn280Ser)). The data were inconclusive for 9 nonsynonymous variants. In Table 4 the numbers of truncating, splice-site, and nonsynonymous mutations are summarized per heart defect diagnosis group.
Discussion
We present the results of NOTCH1 mutation analysis in 428 probands with nonsyndromic congenital LS-CHD. We detected 14 clearly pathogenic NOTCH1 mutations (truncating, RNA splicing, or whole-gene deletion) in 428 patients (3%). The mutation rate was seven times as high in patients with familial compared with nonfamilial LS-CHD. Mutations were present in probands with BAV, AVS, and COA, as well as HLHS. In nonfamilial LS-CHD we detected three mutations (1%), two of which were de novo. NOTCH1 mutations were highly penetrant: 75% of all mutation carriers (probands excluded) showed a cardiovascular abnormality at examination. Nonpenetrance in these families was determined in 12 NOTCH1 mutation carriers (25%), and their mutations were inherited from the mother just as frequently as from the father. The frequency of clearly pathogenic mutations in this cohort of patients with LS-CHD is higher than in previous studies. In five previous studies of NOTCH1 in LS-CHD cohorts, a total of 273 probands were included, and only three clearly pathogenic mutations (truncating or RNA splicing) were reported (1%)15,31,32,33,34: two from a cohort of 53 patients with HLHS33 and one from a cohort of 11 patients with familial BAV.34 In three other studies, only nonsynonymous variants of unknown significance were reported.15,31,32 The higher frequency of NOTCH1 mutations in our population may be due to our sequencing protocols (in one study only four exons were screened),32 or to patient/study population characteristics. Our study population was a selection of patients with LS-CHD who were referred for genetic counseling, and this was reflected in the relatively high percentage of familial cases (35%) compared with other studies.15,31,33
In addition, our population was relatively young, with a median age of 10 years, whereas three of the other studies included mainly adult patients.31,32,34 Congenital heart defects detected in childhood are generally more severe, and this may indicate that NOTCH1 mutations are more often found in severe disease.
The phenotypes of the probands and relatives with pathogenic NOTCH1 mutations in our study included a wide variety of congenital heart defects; in the majority of families with pathogenic mutations, we show that the spectrum of disease involves not only left-sided heart defects but also right-sided heart defects affecting the pulmonary valve, conotruncal disease including pulmonary atresia with intact ventricular septum, Fallot’s tetralogy and truncus arteriosus, and other CHDs, such as anomalous pulmonary venous return, ASD, and VSD. The occurrence of a wider range of defects affecting the conotruncus of the heart in patients with NOTCH1 mutations is in agreement with the reported role of NOTCH signaling in determining the fate of neural crest–derived cells.35,36 Copy-number variations including the NOTCH1 and JAG1 regions were reported in nonsyndromic Fallot’s tetralogy.37 In addition, Fallot’s tetralogy and pulmonary valve disease in Alagille syndrome are caused by mutations in JAG1 and NOTCH2, supporting our findings that NOTCH signaling is involved not only in aortic valve development but also in pulmonary valve development, suggesting a general role in the development of semilunar valves. The finding of left ventricular noncompaction in one patient might be associated with the NOTCH1 mutation because MIB1 mutations, affecting the NOTCH signaling pathway, were previously reported in left ventricular noncompaction.38 The variability we observed in the severity of heart defects within families with pathogenic mutations and the reduced penetrance indicate that the inheritance is not simply monogenic but more complex, and that the phenotype also depends on unknown modifiers. Truncating NOTCH1 mutations were also recently reported in Adams-Oliver syndrome, a genetically heterogeneous syndrome with aplasia cutis and heart malformations in some patients.39,40 The cardiac malformations reported in NOTCH1 mutation carriers among these patients are in the LS-CHD and RS-CHD/CTD spectrum, comparable with the findings in our families. Of note is that no features of Adams-Oliver syndrome were present in our families.
Apart from these clearly pathogenic mutations, which are considered as definitely causing disease, we detected 24 nonsynonymous variants; 6 were novel and 18 were reported in the ExAC database (http://exac.broadinstitute.org) in 35 patients (8%), compared with 5.5% in the five previous studies (corrected for polymorphisms c.3836G>A;p.(Arg1279His) (MAF 0.0231), c.4129C>T;p.(Pro1377Ser) (MAF 0.0234), and c. 6853G>A;p.(Val2285Ile) (MAF 0.0162), which were excluded in our study). One report included functional studies suggesting that two of these variants, present in 6/91 patients, are pathogenic.15 The NOTCH1 gene is large, containing 34 exons, and shows many variants in coding and noncoding regions. These variants may not be causative alone but may well contribute to disease development in a complex model, as reported for variants in other genes involved in congenital heart defects.41 This may be supported by our finding that the allele frequency of five variants—p.(Glu848Lys), p.(Arg912Trp), p.(Gly1091Ser), p.(Thr1344Met), and p.(Arg1350Leu)—detected in more than one family is higher than the allele frequency reported in the Exome Sequencing Project and ExAC databases (Supplementary Table S1 online). We found most nonsynonymous variants in HLHS (16%) and lower frequencies in COA and in BAV/AVS/aortic valve insufficiency (6%). This difference was significant but not reported in an earlier study that also focused on all LS-CHD subgroups.15 Why these variants are more frequent in the most severe LS-CHD group is not clear, but this may indicate that there is a contribution of (some) of these NOTCH1 variants acting in a complex disease model. The pathogenicity of these variants can only be estimated on the basis of software prediction programs and the observed segregation in the family. Therefore, sequencing data from other large cohorts and functional studies are needed to determine the contribution of these variants to congenital heart defects before they can be used in genetic counseling for individual families.
This study has some limitations. Since we only included patients with LS-CHD who were referred for genetic counseling, functional studies were not carried out, synonymous and intronic variants were not analyzed extensively, and mutation analysis of other genes (incidentally) associated with LS-CHD was not performed. However, this is to our knowledge the largest study to date of NOTCH1 in LS-CHD, showing that NOTCH1 mutations play an important role in both LS-CHD and RS-CHD/CTD. Although we detected NOTCH1 mutations in 7% of the familial cases of this cohort, a large number of familial LS-CHD cases remained unsolved, indicating the involvement of other genes associated with a phenotype similar to that presented in the NOTCH1-related families. Massive parallel sequencing will hopefully reveal more about the etiology of LS-CHD and other congenital heart defects in the near future.42
In conclusion, disease-causing NOTCH1 mutations were detected in 7% of familial nonsyndromic LS-CHD and in 1% of sporadic LS-CHD. In addition, we show that the penetrance of heart defects in mutation carriers is high (75%), and the expression of NOTCH1 mutations is variable between and within families, in severity as well as in the cardiac phenotype. Mutations in NOTCH1 are the major disease-causing factors in some families, but additional factors might be involved that modify the phenotype.
We recommend NOTCH1 mutation screening in patients with LS-CHD, pulmonary valve disease, or conotruncal anomalies, especially in familial cases. The finding of a pathogenic NOTCH1 mutation not only helps to counsel the patients at risk for complications such as early calcification of the aortic valve and TAA, it also enables the identification of relatives at risk for complications of a previously unknown congenital heart defect, for instance, BAV with TAA. In addition, it identifies relatives at risk for having affected offspring. Future research is needed to explain the phenotypic variability in NOTCH1 mutations and to reveal other factors involved in familial and sporadic LS-CHD.
Disclosure
The authors declare no conflict of interest.
References
Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol 2004;44:138–143.
McBride KL, Pignatelli R, Lewin M, et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am J Med Genet A 2005;134A:180–186.
Hinton RB Jr, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson DW. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol 2007;50:1590–1595.
Kerstjens-Frederikse WS, Du Marchie Sarvaas GJ, Ruiter JS, et al. Left ventricular outflow tract obstruction: should cardiac screening be offered to first-degree relatives? Heart 2011;97:1228–1232.
Hitz MP, Lemieux-Perreault LP, Marshall C, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet 2012;8:e1002903.
Michelena HI, Khanna AD, Mahoney D, et al. Incidence of aortic complications in patients with bicuspid aortic valves. JAMA 2011;306:1104–1112.
Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol 2010;55:2789–2800.
Hirsch JC, Copeland G, Donohue JE, Kirby RS, Grigorescu V, Gurney JG. Population-based analysis of survival for hypoplastic left heart syndrome. J Pediatr 2011;159:57–63.
Shuhaiber J, Morgan B, Gottliebson W. Survival outcomes following norwood procedure for hypoplastic left heart. Pediatr Cardiol 2015;36:57–63.
Mattina T, Perrotta CS, Grossfeld P. Jacobsen syndrome. Orphanet J Rare Dis 2009;4:9.
Pierpont ME, Basson CT, Benson DW Jr, et al.; American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 2007;115:3015–3038.
Schrander-Stumpel CT, Spruyt L, Curfs LM, Defloor T, Schrander JJ. Kabuki syndrome: clinical data in 20 patients, literature review, and further guidelines for preventive management. Am J Med Genet A 2005;132A:234–243.
Gripp KW, Hopkins E, Jenny K, Thacker D, Salvin J. Cardiac anomalies in Axenfeld-Rieger syndrome due to a novel FOXC1 mutation. Am J Med Genet A 2013;161A:114–119.
Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature 2005;437:270–274.
McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet 2008;17:2886–2893.
Elliott DA, Kirk EP, Yeoh T, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol 2003;41:2072–2076.
Padang R, Bagnall RD, Richmond DR, Bannon PG, Semsarian C. Rare non-synonymous variations in the transcriptional activation domains of GATA5 in bicuspid aortic valve disease. J Mol Cell Cardiol 2012;53:277–281.
Shi LM, Tao JW, Qiu XB, et al. GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int J Mol Med 2014;33:1219–1226.
Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutat Res 2001;479:173–186.
Tan HL, Glen E, Töpf A, et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat 2012;33:720–727.
Theis JL, Zimmermann MT, Evans JM, et al. Recessive MYH6 mutations in hypoplastic left heart with reduced ejection fraction. Circ Cardiovasc Genet 2015;8:564–571.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999;284:770–776.
Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006;7:678–689.
Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci 2013;126:2135–2140.
Bray S, Bernard F. Notch targets and their regulation. Curr Top Dev Biol 2010;92:253–275.
Timmerman LA, Grego-Bessa J, Raya A, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99–115.
Luna-Zurita L, Prados B, Grego-Bessa J, et al. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 2010;120:3493–3507.
Theodoris CV, Li M, White MP, et al. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 2015;160:1072–1086.
Acharya A, Hans CP, Koenig SN, et al. Inhibitory role of Notch1 in calcific aortic valve disease. PLoS One 2011;6:e27743.
Ducharme V, Guauque-Olarte S, Gaudreault N, Pibarot P, Mathieu P, Bossé Y. NOTCH1 genetic variants in patients with tricuspid calcific aortic valve stenosis. J Heart Valve Dis 2013;22:142–149.
Mohamed SA, Aherrahrou Z, Liptau H, et al. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun 2006;345:1460–1465.
McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM 3rd . Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg 2007;134:290–296.
Iascone M, Ciccone R, Galletti L, et al. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin Genet 2012;81:542–554.
Foffa I, Ait Alì L, Panesi P, et al. Sequencing of NOTCH1, GATA5, TGFBR1 and TGFBR2 genes in familial cases of bicuspid aortic valve. BMC Med Genet 2013;14:44.
Wakamatsu Y, Maynard TM, Weston JA. Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development 2000;127:2811–2821.
High FA, Zhang M, Proweller A, et al. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest 2007;117:353–363.
Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 2009;41:931–935.
Luxán G, Casanova JC, Martínez-Poveda B, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med 2013;19:193–201.
Stittrich AB, Lehman A, Bodian DL, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet 2014;95:275–284.
Southgate L, Sukalo M, Karountzos AS, et al. Haploinsufficiency of the NOTCH1 receptor as a cause of Adams-Oliver syndrome with variable cardiac anomalies. Circ Cardiovasc Genet 2015;8:572–581.
Wessels MW, Willems PJ. Genetic factors in non-syndromic congenital heart malformations. Clin Genet 2010;78:103–123.
Pediatric Cardiac Genomics Consortium. The congenital heart disease genetic network study: rationale, design, and early results. Circ Res 2013;112:698–706.
Acknowledgements
The authors thank all patients for their participation in this study, Jackie Senior for editing the manuscript, Tom de Vries Lentsch for preparing the artwork, and Lennart F. Johansson, Krista K. van Dijk-Bos, Esther J. de Jong, Krista A. Kooi, and Ludolf G. Boven for sequencing NOTCH1 and performing the RNA-splicing studies.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Methods
(DOC 25 kb)
Supplementary Table S1
(DOC 162 kb)
Rights and permissions
About this article
Cite this article
Kerstjens-Frederikse, W., van de Laar, I., Vos, Y. et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med 18, 914–923 (2016). https://doi.org/10.1038/gim.2015.193
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.193
Keywords
This article is cited by
-
Genome-wide association studies highlight novel risk loci for septal defects and left-sided congenital heart defects
BMC Genomics (2024)
-
The phenotypic spectrum of terminal 6q deletions based on a large cohort derived from social media and literature: a prominent role for DLL1
Orphanet Journal of Rare Diseases (2023)
-
Concurrent pathogenic variants in SLC6A1/NOTCH1/PRIMPOL genes in a Chinese patient with myoclonic-atonic epilepsy, mild aortic valve stenosis and high myopia
BMC Medical Genetics (2020)
-
Notch and interacting signalling pathways in cardiac development, disease, and regeneration
Nature Reviews Cardiology (2018)
-
Importance of complete phenotyping in prenatal whole exome sequencing
Human Genetics (2018)