Abstract
Purpose:
Recent published studies have demonstrated the incremental value of the use of cell-free DNA for noninvasive prenatal testing with 100% sensitivity for trisomies 21 and 18 and a specificity of ≥99.7% for both. Data presented by two independent groups suggesting positive results by noninvasive prenatal testing were not confirmed by cytogenetic studies.
Methods:
Concordance of results among cases with noninvasive prenatal testing referred for cytogenetic prenatal and/or postnatal studies by karyotyping, fluorescence in situ hybridization, and/or oligo–single-nucleotide polymorphism microarray was evaluated for 109 consecutive specimens.
Results:
Cytogenetic results were positive for trisomy 21 in 38 of the 41 noninvasive prenatal testing–positive cases (true-positive rate: 93%) and for trisomy 18 in 16 of the 25 noninvasive prenatal testing–positive cases (true-positive rate: 64%). The true-positive rate was only 44% (7/16 cases) for trisomy 13 and 38% (6/16 cases) for sex chromosome aneuploidy.
Conclusion:
These findings raise concerns about the limitations of noninvasive prenatal testing and the need for analysis of a larger number of false-positive cases to provide true positive predictive values for noninvasive testing and to search for potential biological or technical causes. Our data suggest the need for a careful interpretation of noninvasive prenatal testing results and cautious transmission of the same to providers and patients.
Genet Med 17 3, 234–236.
Similar content being viewed by others
Main
The evaluation of circulating cell-free DNA by massively parallel shotgun or targeted sequencing to determine the risk of fetal aneuploidy has been rapid and extensive. Recent published studies have demonstrated the incremental value of the use of cell-free DNA for noninvasive prenatal testing (NIPT).1 Various methods and technologies have been used for NIPT, with impressive results. In one study, NIPT demonstrated 100% sensitivity for both trisomy 21 and trisomy 18, with a specificity of ≥99.7% for both.1 Data recently presented by two independent groups in 20132 and 20143 prompted us to review the concordance of results among cases with positive or negative NIPT results referred to Quest Diagnostics for confirmation with cytogenetic studies.
Materials and Methods
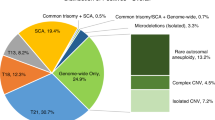
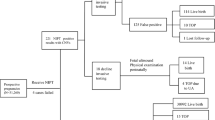
We evaluated the results from 109 consecutive specimens prenatally and/or postnatally studied by standard karyotyping, fluorescence in situ hybridization analysis (AneuVysion; Abbott Molecular/Vysis, Abbott Park, IL), and/or oligo–single-nucleotide polymorphism microarrays (CytoScanHD; Affymetrix, Santa Clara, CA) after NIPT. The NIPT providers were listed in 42 cases and included Panorama (Natera, San Carlos, CA; 20 cases), Harmony (Ariosa Diagnostics, San Jose, CA; 13 cases), MaterniT21 (Sequenom, San Diego, CA; 8 cases), and Verifi (Illumina, Redwood City, CA; 1 case). The most common initial NIPT-positive result was trisomy 21 (41 cases), followed by trisomy 18 (25 cases), trisomy 13 (16 cases), sex chromosome aneuploidy (16 cases), trisomy 16 (3 cases), monosomy 21 (2 cases), and 1 case each of triploidy and microdeletion of 22q11.2. Four samples negative for NIPT but positive for ultrasound findings were included.
Results
Cytogenetic results were positive for trisomy 21 in 38 of the 41 NIPT-positive cases (true-positive rate: 93%) and for trisomy 18 in 16 of the 25 NIPT-positive cases (true-positive rate: 64%) ( Table 1 ). The true-positive rate was only 44% (7/16 cases) for trisomy 13 and 38% (6/16 cases) for sex chromosome aneuploidy. A total of six cases with positive NIPT results for either monosomy 21, trisomy 16, triploidy, or 22q11.2 microdeletion had normal cytogenetic findings. Only one case had very-low-level mosaicism (~5–10%) for trisomy 16. Confined placental mosaicism was confirmed in two cases (2/105, 2%): one with 3% mosaic for trisomy 18 and another with a mosaic segmental uniparental disomy for 11p15.5-p11.2. A false-negative result for NIPT was identified in nonmosaic trisomies 9 and 21, a marker chromosome, and a mosaic sex chromosome aneuploidy.
The findings from our laboratory and those presented by the above-mentioned two groups (n = 80 and n = 46) show that the positive predictive value (PPV) for the three data sources is highest for cases positive for trisomy 21 by NIPT (119/126, 94.4%; Table 2 ). A significantly lower PPV (P < 0.001, χ2 analysis) is displayed for trisomy 18 (25/42, 59.5%), trisomy 13 (12/27, 44.4%), and sex chromosome aneuploidy (11/29, 37.9%). This finding invigorates further thoughts regarding the specificity and PPV of NIPT analyses.
Discussion
The PPV for any test is proportional to the specificity of the assay and the prevalence of the disorder. It is very important to perceive that PPV is not intrinsic to the test; PPV also depends on the prevalence of the condition in the tested population, which was discussed in a recent review of NIPT.4 Thus, the PPV of NIPT is determined not only by the sensitivity and specificity of the assay, but also the prevalence of the tested abnormalities at the gestational age when NIPT is offered (Supplementary Table S1 online).5 For instance, the prevalence of trisomies 21, 18, and 13 in a 35-year-old woman with a fetus at 10 weeks’ gestational age is 1:185, 1:470, and 1:1,500, respectively.5 Assuming a sensitivity and specificity of 99.9% for each abnormality, the PPV would be 84, 68, and 40%, respectively (Supplementary Table S1 online). The observed PPV in this report was 94.4% for trisomy 21, 59.5% for trisomy 18, and 44.4% for trisomy 13. This finding could be due to a higher prevalence of Down syndrome, a higher specificity of NIPT for trisomy 21 than other aneuploidies, or both.
These findings suggest the need for a careful interpretation of NIPT results and cautious transmission of the same to providers and patients. It is vital to educate ordering physicians regarding the differences between specificity and PPV. To an average clinician, the claim that a test is >99% specific leads him or her to expect that the false-positive rate will be <1%. As can been seen by the data in this report and others, the ability of NIPT to correctly predict a positive result for trisomy18 and trisomy 13 is less than 60% ( Table 2 ). For this reason, it is crucial that providers and consumers understand that NIPT is fundamentally a screening test and cannot be used as a replacement for invasive prenatal diagnosis. For a couple with an NIPT positive for trisomy 18, it is foremost to counsel them that the likelihood their fetus actually has a trisomy 18 is <60%. A normal ultrasound result would reduce that likelihood even further. This is very different from being told that there is a 99% chance the fetus is affected and might lead to a different decision regarding an invasive prenatal diagnosis before any action is taken. Clinicians must be educated regarding the differences between published specificities and the more important statistics of PPV.
It is important to note that our data did not differentiate among the various laboratories performing NIPT. There are four main laboratories performing clinical NIPT in the United States, and each uses different methodologies or algorithms. There may be individual differences in specificities that would lead to differing PPV among the various laboratories performing NIPT. A more extensive data set would be required before those calculations could be performed.
In conclusion, use of conventional cytogenetics as the reference standard unravels a rather significant discordance with positive NIPT results. These data should compel us to take a cautious look at NIPT data sets and their sources. As more commercial providers advocate this test, or sponsor academic centers to carry them out, a more diligent comparison of NIPT results with cytogenetic tests should be undertaken.
Disclosure
The authors declare no conflict of interest.
References
Bianchi DW, Parker RL, Wentworth J, et al.; CARE Study Group. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med 2014;370:799–808.
Choy KW, Kwok KY, Lau ET, et al. Discordant karyotype results among non-invasive prenatal screening positive cases. In: The Shifting Landscape of Genetic Testing: Approaches and Success Stories, Platform Session, Abstract #19, American Society of Human Genetics 2013 Annual Meeting, Boston, MA, USA, 2013.
Meck JM, Dugan EK, Aviram A, et al. Non-invasive prenatal screening: a cytogenetic perspective. In: Oral Platform Presentations: Cytogenetics, Abstract #17, American College of Medical Genetics and Genomics 2014 Annual Meeting, Nashville, Tennessee, USA, 2014.
Lutgendorf MA, Stoll KA, Knutzen DM, Foglia LM . Noninvasive prenatal testing: limitations and unanswered questions. Genet Med 2014;16:281–285.
Gardner RJ, Sutherland GR, Shaffer LG . Chromosome Abnormalities and Genetic Counseling. 4th edn. Oxford University Press: New York, 2012.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1
(DOC 64 kb)
Rights and permissions
About this article
Cite this article
Wang, JC., Sahoo, T., Schonberg, S. et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genet Med 17, 234–236 (2015). https://doi.org/10.1038/gim.2014.92
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.92
Keywords
This article is cited by
-
Positive predictive value of noninvasive prenatal testing for sex chromosome abnormalities
Molecular Biology Reports (2022)
-
Nicht invasive Pränataldiagnostik mittels molekulargenetischer Tests (NIPT) zur Erkennung der Trisomien 13, 18 und 21
Medizinische Genetik (2019)
-
Performance of non-invasive prenatal testing for trisomies 21 and 18 in twin pregnancies
Molecular Cytogenetics (2018)
-
The application of NIPT using combinatorial probe-anchor synthesis to identify sex chromosomal aneuploidies (SCAs) in a cohort of 570 pregnancies
Molecular Cytogenetics (2018)
-
Developing context-specific next-generation sequencing policy
Nature Biotechnology (2016)