Abstract
Purpose:
Hirschsprung disease (OMIM 142623) is a neurocristopathy attributed to a failure of cell proliferation or migration and/or failure of the enteric precursors along the gut to differentiate during embryonic development. Although some genes involved in this pathology are well characterized, many aspects remain poorly understood. In this study, we aimed to identify novel genes implicated in the pathogenesis of Hirschsprung disease.
Methods:
We compared the expression patterns of genes involved in human stem cell pluripotency between enteric precursors from controls and Hirschsprung disease patients. We further evaluated the role of DNMT3B in the context of Hirschsprung disease by inmunocytochemistry, global DNA methylation assays, and mutational screening.
Results:
Seven differentially expressed genes were identified. We focused on DNMT3B, which encodes a DNA methyltransferase that performs de novo DNA methylation during embryonic development. DNMT3B mutational analysis in our Hirschsprung disease series revealed the presence of potentially pathogenic mutations (p.Gly25Arg, p.Arg190Cys, and p.Gly198Trp).
Conclusion:
DNMT3B may be regulating enteric nervous system development through DNA methylation in the neural crest cells, suggesting that aberrant methylation patterns could have a relevant role in Hirschsprung disease. Moreover, the synergistic effect of mutations in both DNMT3B and other Hirschsprung disease–related genes may be contributing to a more severe phenotype in our Hirschsprung disease patients.
Genet Med 16 9, 703–710.
Similar content being viewed by others
Introduction
Hirschsprung disease (HSCR; OMIM 142623) is the most common neurocristopathy in humans, affecting 1:5,000 newborns. It appears either sporadically or on a familial basis and may be associated with other developmental defects. HSCR results from a failure to fully colonize the gut by neural crest cells (NCCs) of the enteric nervous system (ENS). This failure results in the absence of enteric ganglia in a variable segment of the distal bowel and causes severe intestinal dysfunction. HSCR phenotype can be classified into two main groups: short-segment forms (S-HSCR), which include patients with aganglionosis as far as the splenic flexure, and long-segment forms (L-HSCR), in which aganglionosis extends beyond the splenic flexure.1,2
The RET proto-oncogene (OMIM 164761) is the main gene associated with HSCR, although other genes (GDNF, NRTN, PSPN, EDNRB, EDN3, ECE1, NTF3, NTRK3, SOX10, PHOX2B, L1CAM, ZFHX1B, KIAA1279, TCF4, PROK2, PROKR1, PROKR2, GFRA1, NRG1, NRG3, SEMA 3A, and SEMA 3D) have been also related to the disease.2,3,4,5,6,7,8,9,10,11 HSCR is regarded as a disorder with complex genetic basis. The contribution of different mutational events within several loci acting in an additive manner is usually involved in the development of this disease.2,11 This genetic complexity observed in HSCR could be explained in part by the nature of the development of the ENS, which is regulated by an ever-increasing range of molecules and signaling pathways involving both NCCs and the intestinal environment.12,13
To elucidate the genetic basis of HSCR, one must understand the mechanisms underlying the selective gene expression during the development of the ENS. In cells, all genes are transcribed at some stage of the life cycle, and the selection of genes to be activated or specifically inactivated plays a key role for proper ENS formation. One of the main regulatory processes implicated in the maintenance of the correct levels of gene expression is DNA methylation–dependent transcriptional silencing. For instance, failure to appropriately repress genes has been related to many human diseases, including neurodevelopmental disorders and cancer.14,15 DNA methylation patterns are established by three independent DNA methyltransferases—DNMT1, DNMT3A, and DNMT3B16,17—known to be necessary during neural progenitor cell development.18,19,20 DNMT1 is mainly implicated in keeping specific methylation during DNA replication, whereas DNMT3A and DNMT3B contribute to the establishment of de novo methylation patterns during early embryogenesis. In addition, interaction of DNMT3A and DNMT3B has also been observed in de novo methylation processes.21 Specifically, DNMT3B has an important role in the initial steps of progenitor cell differentiation, and DNMT3A is essential for the maturation processes of cell differentiation.15 This de novo DNA methylation of the genome suggests the existence of conserved mechanisms during mammalian development.22,23,24
Nowadays, the isolation of ENS progenitor cells from the human postnatal gut represents a powerful tool for the study of ENS development. The ENS progenitor cells are cultured in order to form cell clusters, which subsequently grow as aggregates or neurosphere-like bodies (NLBs) in suspension and include stem cells and their progeny derived from the neural crest. These NLBs have been used as a source of enteric precursor cells that can be transplanted to restore the contractile properties of aganglionic bowel by neurally mediated mechanisms.25,26,27,28
In the present study, we have used such NLBs to search for new genes that could be implicated in ENS formation and in the pathogenesis of HSCR. Given that neural crest pluripotency is essential for proper ENS development in particular, we have studied the expression profiles of pluripotency-associated genes in enteric neural precursors. We performed a comparative study in NLBs derived from the postnatal human bowels of a selection of HSCR patients (HSCR-NLBs) versus controls (control-NLBs). Among the differentially expressed genes, we have identified the de novo DNA methyltransferase DNMT3B, which plays an important role during the early stages of genome-wide DNA methylation and is highly implicated in neural progenitor differentiation during ENS development. Moreover, due to its important role in shaping DNA methylation patterns, we carried out a mutational screening in our series of HSCR patients to determine if mutations in DNMT3B might also be associated with HSCR.
Materials and Methods
Ethical approval
Written informed consent for surgery and clinical and molecular genetic studies was obtained from all participants. The study was approved by the ethics committee for clinical research of the University Hospital Virgen del Rocío (Seville, Spain) and complies with the tenets of the Declaration of Helsinki.
Generation of ENS human NLBs
Human ENS progenitor cells were isolated from human postnatal tissues of ganglionic full-thickness gut of seven sporadic nonrelated patients diagnosed with isolated HSCR (L-HSCR:S-HSCR = 4:3; males:females = 4:3) and four patients without HSCR considered as controls (two males and two females). The age range of the subjects studied here was 3 months to 3 years. None of the seven HSCR patients carried coding mutations in RET or other HSCR-related genes such as GDNF, EDN3, EDNRB, SOX10, or PHOX2B. Regarding the presence of the RET enhancer mutation c.73+9277T>C (rs2435357), which has been described as being significantly associated with HSCR susceptibility, making a 20-fold greater contribution to risk than coding mutations,11 the three possible genotypic combinations were represented in our patients (one patient CC, three patients CT, and three patients TT). The ganglionic bowels were dissected and incubated in a solution of 0.26 mg/ml trypsin collagenase, 5 mg/ml dispase, 0.28 mg/ml hyaluronidase, 3.3 μg/ml elastase, and 0.6 mg/ml collagenase in phosphate-buffered saline for up to 30 minutes at 37 °C. Digested tissue was triturated and washed, and the cells were cultured in Dulbecco’s modified Eagle medium (1 mg/ml glucose) containing 100 U/ml penicillin, 100 g/ml streptomycin, supplemented with 2 mmol/l l-glutamine (Gibco Life Technologies, Carlsbad, CA), 0.05 mmol/l 2-mercaptoethanol, 1% (volume/volume) N1 (Sigma Aldrich, Poole, Dorset, UK), 10% (volume/volume) human serum, 20 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor, and 10 ng/ml glial cell–derived neurotrophic factor (Peprotech, London, UK), as previously described by Ruiz-Ferrer et al.7
Immunocytochemical analysis showed that the majority of the ENS progenitors in our culture were nestin-positive cells. To characterize the cells that form the NLBs, we proceeded to adhere them, and subsequently they were immunostained with a neuronal marker (TuJ1), glial markers (glial fibrillary acidic protein, S100), and a smooth muscle alpha marker. RET was also expressed by NLBs, and confocal analysis revealed the presence of this receptor in all nestin-positive NLBs, as previously described by Ruiz-Ferrer et al.7
DNA isolation from neural precursor cells (NLBs)
Frozen NLBs from controls and HSCR patients were digested with proteinase K (0.02 mg/ml) in the presence of lysis buffer (Tris, 20 mmol/l (pH 7.8); NaCl, 150 mmol/l; Igepal, 1%; and EDTA, 2 mmol/l) at 37 °C overnight, followed by phenol/chloroform/isoamilic extraction and ethanol precipitation and finally dissolved in 20 or 50 µl of high–molecular-grade water.
Gene expression analysis by quantitative real-time polymerase chain reaction
For the characterization of enteric precursor cells, we performed an expression study of genes previously described as markers of stem cells. We considered the pluripotency array as the one that better fits to characterize our cultures, due to the lack of well-defined expression patterns in human postnatal enteric precursors. Purification of messenger RNA (mRNA) and synthesis of complementary DNA from neural precursor cells of NLBs as well as from ganglionic gut tissues from HSCR patients and controls were performed using the protocol provided by the µMACS mRNA isolation Kit and the µMACS cDNA Synthesis Kit (MACS Molecular, Germany) in a thermoMAKS Separator (MACS Miltenyi Biotech, Germany). The expression study was made using the 7900HT Fast Real-Time PCR system (Life Technologies) with TaqMan Array Gene Signature Plates (TaqMan Human Stem Cell Pluripotency Array, Life Technologies). The results were analyzed using the RQ Manager Software (Life Technologies), establishing glyceraldehyde-3-phosphate dehydrogenase as our endogenous control. Following the software recommendations, the upper limit of the cycle threshold (Ct) was 32, and therefore we considered positive expression exclusively when Ct values were <32. The same protocol was carried out for the expression analysis of DNMT3B in ganglionic gut tissue from HSCR patients and controls.
Global DNA methylation assays
The global DNA methylation levels in neural precursor cells from the control-NLBs and HSCR-NLBs were measured with an enzyme-linked immunosorbent assay–based commercial kit (MDQ1, Imprint Methylated DNA Quantification Kit, Sigma Aldrich, St Louis, MO). For this assay, we used 100 ng of DNA, which was diluted in the corresponding volume of lysis and binding buffers and incubated at 37 °C. The methylated fraction of DNA was recognized by 5-methylcytosine antibody and quantified through an enzyme-linked immunosorbent assay reaction (450 nm). Quantification of global DNA methylation was obtained from calculating the amount of 5-methylcytosine in control-NLBs and HSCR-NLBs relative to global cytidine (5-methylcytosine + deoxycytidine) in a positive control that had been methylated previously. All the samples were analyzed in triplicate.
Statistical analyses for gene expression and global DNA methylation
Data are presented as the mean ± SEM of values obtained from at least three experiments. Comparisons between values obtained in control-NLBs and HSCR-NLBs were analyzed using the Student’s t-test. Differences were considered significant when the P value was ≤ 0.05.
Inmunocytochemistry
Immunocytochemical studies were intended to observe the DNMT3B expression in NLBs. The single cells from NLBs were seeded onto fibronectin-poly d lysine–coated coverslips and fixed with 4% (weight/volume) paraformaldehyde in 0.1 mol/l phosphate-buffered saline. Then, they were incubated for 1 hour in 2.5% (weight/volume) bovine serum albumin in phosphate-buffered saline and with primary and secondary antibodies. After washing, the coverslips were mounted on slides with Fluoro-Gel (EMS, Hatfield, PA), and fluorescent signals were detected using a Leica Spectra confocal microscope (Leica Microsystem, Wetzlar, Germany). The primary antibody used was anti-DNMT3B (rabbit polyclonal 1:100; Santa Cruz Biotechnology, Dallas, TX). The secondary antibody used was antirabbit immunoglobulin G labeled with Alexa Fluor 568 (1:200; Life Technologies), as previously described by Ruiz-Ferrer et al.6 Omission of primary antibodies resulted in no detectable staining in all cases.
Mutational analysis of DNMT3B
Mutational study of DNMT3B was performed in our series of HSCR patients and a group of controls. We included a total of 170 Spanish HSCR patients, and their parents when available. One hundred and sixty-eight were sporadic cases, whereas two were familial cases belonging to the same family. In addition, we also analyzed a group of 150 normal controls comprising unselected, unrelated, race-, age-, and sex-matched individuals.
Genomic DNA was extracted from peripheral blood leukocytes from all individuals included in the study, using standard protocols.
The mutational screening of the complete coding sequence of DNMT3B (NM_006892.3) was carried out by direct sequencing using an ABI PrismH3730 Genetic Analyzer and the SeqScapeH v2.5 software (Life Technologies, Carlsbad, CA). Primers used for polymerase chain reaction amplification and direct sequencing, as well as the annealing temperatures used, are summarized in Supplementary Table S1 online.
The novel identified variants were tested in a group of 150 healthy control subjects to determine if the variant was a common polymorphism. Moreover, segregation analysis of the identified variants was performed when DNA samples were available.
To evaluate the pathogenicity of the novel variants at the protein sequence level, we used different bioinformatics tools. We have analyzed the potential impact of the amino acid substitution on the structure or function of the human encoded protein using the SIFT and Polyphen tools (http://sift.bii.a-star.edu.sg/ and http://genetics.bwh.harvard.edu/pph/). Possible occurrence in regulatory or splicing motifs of novel variants located within the noncoding region were submitted to the Splice Sites and Transcription Factors Binding sequences prediction interfaces (http://www.fruitfly.org/seq_tools/splice.html, http://www.fruitfly.org/seq_tools/promoter.html, and http://http://alggen.lsi.upc.es/cgi-bin/promo-v3/promo/promainit.cgi?dirDB=TE8.3). The DNMT3B protein sequence was submitted to ScanProsite (http://expasy.org/tools/scanprosite/) to study predicted domains of the protein, profiles, and motifs affected by the identified variants.
Finally, all the variants detected were checked in Ensembl (http://www.ensembl.org/), NCBi (http://www.ncbi.nlm.nih.gov/), HGMD (http://www.hgmd.org/), and the Exome variant server (http://evs.gs.washington.edu/EVS/).
Results
Analysis of differential expression of pluripotency stem cell markers in NLBs from HSCR patients versus controls
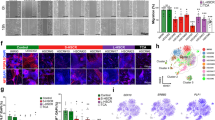
For the characterization of HSCR neural precursor cells of the ENS, we analyzed the gene expression profiles of enteric precursor cells using a TaqMan Human Stem Cell Pluripotency Array on HSCR-NLBs and control-NLBs. We observed positive expression for 24 pluripotency stem cell markers (see Supplementary Table S2 online) of the 96 genes analyzed (see Supplementary Table S3 online). This study showed that seven genes presented statistically significant differential expression (>twofold change) between HSCR-NLBs and control-NLBs (three upregulated and four downregulated; P value ≤ 0.05). These genes include five implicated in cell proliferation and migration (NESTIN, FN1, LAMC1, PECAM, and SEMA3A), one encoding for a transcription factor (PAX6), and one encoding for a de novo DNA methyltransferase (DNMT3B) ( Table 1 and Figure 1 ). Our results showed a higher expression of NESTIN, PECAM, and SEMA3A mRNAs in HSCR-NLBs as compared with control-NLBs and downregulation of FN1, LAMC1, and PAX6 mRNAs, as well as DNMT3B mRNA. The same expression patterns of genes such as SEMA3A and XIST have been previously described,29,30 which allowed us to validate the results obtained in this study.
Genes with a significant differential expression in HSCR-NLBs versus control-NLBs. Heat maps were generated using DataAssist v3.0 software (Life Technologies) and represent the messenger RNA expression levels of seven genes that have a differential expression profile between (a) HSCR-NLBs and control-NLBs and (b) colon tissue from HSCR patients and that from controls. Genes were hierarchically clustered by Pearson correlation coefficient using average linkage. The color scale, representing ΔCt, is shown on the right side. Green indicates genes with relatively decreased expression, whereas red indicates genes with relatively increased expression. Ct, cycle threshold; HSCR, Hirschsprung disease; NLBs, neurosphere-like bodies.
On the basis of these results, we focused on DNMT3B for several reasons: first, in all HSCR-NLBs, DNMT3B mRNA was found to be downregulated in comparison with control-NLBs (HSCR-NLBs Ct = 36.8; control-NLBs Ct = 29). These values indicated that the levels of DNMT3B mRNA were reduced by 3.6-fold in HSCR-NLBs (P = 0.0006; Figure 2a ). By contrast, DNMT3B expression was not observed in gut tissue samples from HSCR patients or controls ( Figure 1 ). Second, the decrease in DNMT3B mRNA levels observed in HSCR-NLBs highlights the possibility that there might be variations in DNA methylation patterns between enteric neural precursor cells from HSCR patients and control individuals. Finally, DNMT3B is essential for the initial steps of precursor cell differentiation during embryonic development through de novo DNA methylation. This fact may be crucial in the establishment of cellular identity and lineage choice.
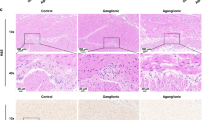
Expression of DNMT3B in HSCR-NLBs as compared with control-NLBs. (a) Graph showing the lower expression of DNMT3B messenger RNA. Data are represented as mean ± SEM. P = 0.0006; confocal images of protein expression of DNMT3B in neural precursors derived from (b) HSCR-NLBs and (c) control-NLBs. Inmunostaining for DNMT3B is shown in red, whereas the cell nucleus was counterstained with 4′,6-diamidino-2-phenylindole (blue). Bar = 25 µm. ENS, enteric nervous system; HSCR, Hirschsprung disease; NLBs, neurosphere-like bodies.
HSCR samples show fewer DNMT3B-positive neural precursors than controls
To further characterize the observed decrease of DNMT3B mRNA levels in HSCR-NLBs, we studied the cellular distribution and protein expression of DNMT3B mRNA in neural precursors derived from control-NLBs and HSCR-NLBs using inmunocytochemistry. We observed that DNMT3B was localized mainly in the cell nucleus, and interestingly, we noticed that the number of positive cells in HSCR-NLBs ( Figure 2b ) was significantly lower than that of control-NLBs ( Figure 2c ). These results are in agreement with our data showing that HSCR-NLBs include precursor cells, which present a different methylation pattern.
Enteric precursor cells derived from HSCR-NLBs showed a decrease in methylation compared with control-NLBs
Considering the reduced DNMT3B mRNA levels in HSCR-NLBs, we performed a study to determine whole-genome methylation levels in our NLB samples. To this purpose, we carried out an enzyme-linked immunosorbent assay–based methylated DNA quantification assay, obtaining a global genomic hypomethylation of 69 ± 15.8% in HSCR enteric precursors as compared with control precursors (100%) (see Supplementary Figure S1 online). These results correlate with the reduced DNMT3B mRNA expression levels found in HSCR-NLBs and are consistent with a potential role of DNMT3B in this pathology.
Identification of DNMT3B mutations in our cohort of HSCR patients
With the aim of clarifying the contribution of DNMT3B to the HSCR phenotype and defining the mutational status of our series of HSCR patients, we performed a mutational screening by direct sequencing of all DNMT3B exons, including intron/exon boundaries.
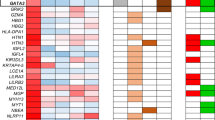
We identified a total of 41 variants in DNMT3B in our HSCR series, 9 of them located within the coding region of the gene, whereas 32 were in the noncoding region (see Supplementary Table S4 online). Bioinformatic analyses of those 32 noncoding variations failed to detect any effect on either the splicing processes or the generation/modification of a transcription factor binding site along the DNA sequence. Regarding the nine coding variants, four were synonymous amino acid substitutions (p.Gly15Gly, p.Cys524Cys, p.Tyr624Tyr, and p.Asp627Asp), and another two (p.Arg42Cys and p.Ala384Thr), although nonsynonymous, were expected to be unlikely to affect the protein structure and function after submitting them to physical and comparative considerations analysis (http://genetics.bwh.harvard.edu/pph/). More interesting was the finding of the remaining three coding variants, predicted to be probably damaging: p.Gly25Arg (rs151128145, minor allele frequency (MAF): 0.001/1), p.Arg190Cys (rs140395707, MAF: not available), and p.Gly198Trp (rs61758433, MAF: 0.001/2). Of note, none of them was detected in our control group ( Figure 3 ).
The p.Gly25Arg mutation was detected in two different nonrelated patients with L-HSCR phenotype (HSCR-1 and HSCR-2). It is worth mentioning that both patients carried other mutations in genes associated with HSCR. Specifically, HSCR-1 also carried a RET mutation (p. Ser147del), which was inherited from his healthy father, but did not carry any pathogenic variant in DNMT3B. A cousin of the proband displaying the S-HSCR phenotype carried the same mutation in RET (p.Ser147del), inherited from his healthy mother, without a pathogenic variant in DNMT3B ( Table 2 ).
The second case, HSCR-2, with the p.Gly25Arg change in DNMT3B, is a L-HSCR female also carrying the p.Gly198Trp mutation in this gene. In addition, she also carries the NTF3 p.Glu76Arg mutation previously reported to be involved in the pathogenesis of HSCR.4 Although the NTF3 mutation was confirmed to be inherited from her healthy mother, the unavailability of DNA from the father of the proband did not allow us to determine the inheritance of both DNMT3B mutations. Nevertheless, given that such DNMT3B mutations were absent in the mother, they were most probably inherited from her father, which leads us to assume that they were located in the same chromosome (in cis) ( Table 2 ).
Finally, the DNMT3B p.Arg190Cys mutation was detected in a male HSCR patient (HSCR-3) with a L-HSCR phenotype, and we verified that the mutation was inherited from his healthy mother.
Discussion
HSCR is a complex neurodevelopmental disorder in which progenitor cells derived from NCCs have deficient cell proliferation, migration, and/or differentiation patterns. Experimental approaches in different animal models using DNA microarray techniques to profile gene expression in the developing ENS31,32 have allowed researchers to better understand the cellular and molecular bases of ENS development.33,34 However, new approaches are still needed for studying the early stages of neurogenesis in the human embryo. Human enteric precursors have become a powerful tool for studying early embryonic stages of gut development.25,26,27,28
In this study, we have evaluated the differences in the expression patterns of pluripotency stem cell marker genes between precursors derived from HSCR-NLBs and control-NLBs. We have obtained different expression levels in a group of genes (NESTIN, FN1, LAMC1, PECAM, SEMA3A, PAX6, and DNMT3B) between the two types of samples. It is worth mentioning that the differential expression of genes such as SEMA3A and XIST was essential for validating these results. In the case of SEMA3A, it has been demonstrated that HSCR colon displays a higher expression of SEMA3A compared with controls.29 Importantly, we have obtained the same results in NLBs derived from human postnatal gut. Furthermore, XIST (X (inactive)-specific transcript), expressed exclusively from the XIC of the inactive X chromosome and essential for initiation and spreading of X inactivation,30 was observed exclusively in NLBs from female HSCR patients and controls. Most of the identified genes encode proteins that are related to cell proliferation or migration, critical processes for proper ENS or central nervous system development.
We have focused on DNMT3B (de novo DNA methyltransferase) due to the importance of epigenetic and de novo methylation events during the selective expression of genes during development, as well as the important role of this methyltransferase in the initial steps of progenitor cell differentiation.15,22,23
We observed reduced levels of DNMT3B mRNA and protein levels in neural precursor cells of HSCR-NLBs. We also detected a high number of enteric precursors with different DNA methylation patterns in HSCR-NLBs, which may lead to variations in the transcriptional regulation and the pluripotency stage during the cell differentiation process of these precursors. The DNA methylation status is highly related to expression levels of methyltansferases, and proper DNA methylation patterns are essential for preserving a correct embryonic development. Watanabe et al.15 reported that DNMT3B is essential for the initial steps in the cell differentiation of enteric precursor cells to neurons in mice. In this sense, we could hypothesize that these changes may induce a transient proliferation arrest in an undifferentiated precursor state, leading to a defective differentiation into the cell lineage required for an appropriate human ENS development. In agreement with these results, we have observed an increase in the stem cell and neural precursor marker NESTIN expression levels in HSCR-NLBs, suggesting that the enteric precursors derived from HSCR-NLBs are in a less differentiated state as compared with control-NLBs.
NCCs are a population of multipotent cells that migrate extensively throughout vertebrate embryos to form diverse structures. The migratory NCCs flow occurs with neuronal differentiation in order to achieve a proper ENS development. Although DNMT3B has been reported to be inessential for the development of mouse cranial NCCs,34 it seems to have a role in the neurogenesis of enteric NCCs. DNMT3B is detected within a narrow window during early neurogenesis, whereas DNMT3A is present in both embryonic and postnatal central nervous system tissue.35 This is probably the reason why we did not observe DNMT3B expression in gut tissues from either HSCR patients or controls.
Methylation levels in HSCR-NLBs were lower compared with those of control-NLBs, which reinforces the hypothesis that enteric precursors of HSCR-NLBs lacking DNMT3B show differential DNA methylation patterns, as well as different expression of genes and cellular identity. We have also identified in enteric precursor cells from HSCR-NLBs significantly downregulated genes implicated in the above-mentioned processes including FN1, LAMC1, and PAX6. Furthermore, hypomethylation of the transcription factor PAX6 results in decreased mRNA expression and reduced proliferation capacity,36,37 and PAX6 overexpression results in a higher neuronal differentiation.38,39
In addition, we have performed the first comprehensive mutational spectrum analysis of DNMT3B in the context of HSCR, which revealed the presence of three potentially pathogenic sequence changes (p.Gly25Arg, p.Arg190Cys, and p.Gly198Trp) in three different patients. The low MAF values reported and the absence of the three variants in our group of normal controls discard them as common polymorphisms and fits with the low incidence of HSCR. It is of interest to note that the three mutations are located in a region of the protein that interacts with DNMT1 to perform the establishment of a de novo methylation pattern that occurs during embryonic development.25 Therefore, these mutations may hamper the appropriate methylation patterns and the regulation of gene expression required for proper embryonic development. The failure in the regulation of gene expression during the development of the ENS could be one of the mechanisms triggering HSCR in these patients. This is consistent with our results from the expression studies, supporting the hypothesis that DNMT3B is a gene involved in the development of ENS and therefore in the occurrence of HSCR. Worth mentioning is the fact that, in at least two cases, the DNMT3B mutations cosegregated with mutations in other HSCR loci, in accordance with a complex and multigenic model of inheritance previously proposed for HSCR. It is currently accepted that both common “low penetrant” variants in combination with rare or private “high-penetrant” variants determine the risk for HSCR. Moreover, length of aganglionosis is suspected to be the result of the contribution of a number of molecular events affecting the proliferation, migration, and/or differentiation of cells during ENS formation: the higher the number of variations in key genes for ENS development and the more deleterious effect of such mutations would lead to an earlier arrest of migration and therefore to a longer distal aganglionic intestinal portion. This would explain the finding of multiplex HSCR families with affected members showing a different phenotype as the result of the contribution of at least one high-penetrant mutation and additional modifying genetic factors. For instance, in the case of a family previously reported by our group, both RET and NTRK3 mutations acting together were necessary and sufficient for the appearance of the disease, and an EDN3 mutation was acting as a phenotype-modifying factor.40Translating this to our current results, we can also propose a synergistic effect of mutations in DNMT3B and other HSCR-related genes, mainly RET, to generate the phenotype. This is particularly interesting for the multiplex family reported here, in which the RET p.Ser147del mutation is present in the two affected members studied (the L-HSCR proband and his S-HSCR cousin). The DNMT3B Gly25Arg mutation appears exclusively in the patient with the L-HSCR, suggesting that the DNMT3B variant may be acting as a phenotype modifier. It is tempting to speculate that the RET mutation may be the major cause of the disease in the family, although—as is the case with the vast majority of RET mutations—with incomplete penetrance,2 whereas the DNMT3B rare variant would contribute a more subtle effect. Regarding the second case, only the NTF3, but not the DNMT3B, mutation was inherited from the healthy mother, again supporting the additive model.
In summary, we have characterized the precursors of human ENS and have identified a group of genes with differential expression patterns between ENS precursors from HSCR patients and controls. Through the experiments designed to study the role of DNMT3B in HSCR, we have observed a decrease of DNA methylation in enteric precursors from HSCR-NLBs, resulting in lower pluripotency. In this study, we have shown for the first time that DNMT3B could be involved in the ENS development, and mutations in its coding sequence may contribute in an additive or multiplicative manner to the severity of the HSCR phenotype. Moreover, further studies are necessary to investigate the role of DNMT3B in the regulation—through DNA methylation—of known genes implicated in this pathology.
Disclosure
The authors declare no conflict of interest.
References
Chakravarti A, Lyonnet S . Hirschsprung disease. In: Scriver CR, Beaudet AL, Valle D, Sly WS, Childs B, Kinzler K, Volgestein B (eds). Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, 2001:6231–6255.
Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al.; Hirschsprung Disease Consortium. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet 2008;45:1–14.
Borrego S, Fernández RM, Dziema H, et al. Investigation of germline GFRA4 mutations and evaluation of the involvement of GFRA1, GFRA2, GFRA3, and GFRA4 sequence variants in Hirschsprung disease. J Med Genet 2003;40:e18.
Ruiz-Ferrer M, Fernandez RM, Antiñolo G, Lopez-Alonso M, Borrego S . NTF-3, a gene involved in the enteric nervous system development, as a candidate gene for Hirschsprung disease. J Pediatr Surg 2008;43:1308–1311.
Fernández RM, Sánchez-Mejías A, Mena MD, et al. A novel point variant in NTRK3, R645C, suggests a role of this gene in the pathogenesis of Hirschsprung disease. Ann Hum Genet 2009;73:19–25.
Ruiz-Ferrer M, Torroglosa A, Luzón-Toro B, et al. Novel mutations at RET ligand genes preventing receptor activation are associated to Hirschsprung’s disease. J Mol Med 2011;89:471–480.
Ruiz-Ferrer M, Torroglosa A, Núñez-Torres R, de Agustín JC, Antiñolo G, Borrego S . Expression of PROKR1 and PROKR2 in human enteric neural precursor cells and identification of sequence variants suggest a role in HSCR. PLoS ONE 2011;6:e23475.
Tang CS, Ngan ES, Tang WK, et al. Mutations in the NRG1 gene are associated with Hirschsprung disease. Hum Genet 2012;131:67–76.
Tang CS, Cheng G, So MT, et al. Genome-wide copy number analysis uncovers a new HSCR gene: NRG3. PLoS Genet 2012;8:e1002687.
Luzón-Toro B, Fernández RM, Torroglosa A, et al. Mutational spectrum of semaphorin 3A and semaphorin 3D genes in Spanish Hirschsprung patients. PLoS ONE 2013;8:e54800.
Emison ES, Garcia-Barcelo M, Grice EA, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet 2010;87:60–74.
Burns AJ, Douarin NM . The sacral neural crest contributes neurons and glia to the post-umbilical gut: spatiotemporal analysis of the development of the enteric nervous system. Development 1998;125:4335–4347.
Kruger GM, Mosher JT, Tsai YH, et al. Temporally distinct requirements for endothelin receptor B in the generation and migration of gut neural crest stem cells. Neuron 2003;40:917–929.
Robertson KD, Wolffe AP . DNA methylation in health and disease. Nat Rev Genet 2000;1:11–19.
Watanabe D, Uchiyama K, Hanaoka K . Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience 2006;142:727–737.
Yen RW, Vertino PM, Nelkin BD, et al. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res 1992;20:2287–2291.
Xie S, Wang Z, Okano M, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 1999;236:87–95.
Okano M, Bell DW, Haber DA, Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257.
Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004;429:900–903.
Oka M, Rodic N, Graddy J, Chang LJ, Terada N . CpG sites preferentially methylated by Dnmt3a in vivo. J Biol Chem 2006;281:9901–9908.
Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S . Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J 2002;21:4183–4195.
Singh RP, Shiue K, Schomberg D, Zhou FC . Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant 2009;18:1197–1211.
Sundberg M, Andersson PH, Åkesson E, et al. Markers of pluripotency and differentiation in human neural precursor cells derived from embryonic stem cells and CNS tissue. Cell Transplant 2011;20:177–191.
Schneider L, d’Adda di Fagagna F . Neural stem cells exposed to BrdU lose their global DNA methylation and undergo astrocytic differentiation. Nucleic Acids Res 2012;40:5332–5342.
Rauch U, Hänsgen A, Hagl C, Holland-Cunz S, Schäfer KH . Isolation and cultivation of neuronal precursor cells from the developing human enteric nervous system as a tool for cell therapy in dysganglionosis. Int J Colorectal Dis 2006;21:554–559.
Almond S, Lindley RM, Kenny SE, Connell MG, Edgar DH . Characterisation and transplantation of enteric nervous system progenitor cells. Gut 2007;56:489–496.
Richard M, Lindley RM, Hawcutt DB, et al. Human and mouse enteric nervous system neurosphere transplants regulate the function of aganglionic embryonic distal colon. Gastroenterology 2008;133:205–216.
Richard M, Lindley RM, Hawcutt DB, Connell MG, Edgar DH, Kenny SE . Properties of secondary and tertiary human enteric nervous system neurospheres. J Pediatr Surg 2009;44:149–156.
Wang LL, Fan Y, Zhou FH, et al. Semaphorin 3A expression in the colon of Hirschsprung disease. Birth Defects Res Part A Clin Mol Teratol 2011;91:842–847.
Pontier DB, Gribnau J . Xist regulation and function explored. Hum Genet 2011;130:223–236.
Heanue TA, Pachnis V . Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc Natl Acad Sci USA 2006;103:6919–6924.
Vohra BP, Tsuji K, Nagashimada M, et al. Differential gene expression and functional analysis implicate novel mechanisms in enteric nervous system precursor migration and neuritogenesis. Dev Biol 2006;298:259–271.
Goldstein AM, Hofstra RM, Burns AJ . Building a brain in the gut: development of the enteric nervous system. Clin Genet 2013;83:307–316.
Jacques-Fricke BT, Roffers-Agarwal J, Gammill LS . DNA methyltransferase 3b is dispensable for mouse neural crest development. PLoS ONE 2012;7:e47794.
Feng J, Chang H, Li E, Fan G . Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res 2005;79:734–746.
Wu X, Rauch TA, Zhong X, et al. CpG island hypermethylation in human astrocytomas. Cancer Res 2010;70:2718–2727.
Balmer NV, Weng MK, Zimmer B, et al. Epigenetic changes and disturbed neural development in a human embryonic stem cell-based model relating to the fetal valproate syndrome. Hum Mol Genet 2012;21:4104–4114.
Jang ES, Goldman JE . Pax6 expression is sufficient to induce a neurogenic fate in glial progenitors of the neonatal subventricular zone. PLoS ONE 2011;6:e20894.
Scardigli R, Bäumer N, Gruss P, Guillemot F, Le Roux I . Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 2003;130:3269–3281.
Sánchez-Mejías A, Fernández RM, López-Alonso M, Antiñolo G, Borrego S . Contribution of RET, NTRK3 and EDN3 to the expression of Hirschsprung disease in a multiplex family. J Med Genet 2009;46:862–864.
Acknowledgements
This work was supported by the Instituto de Salud Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness, Spain (PI1001290), and the Regional Ministry of Innovation, Science and Enterprise of the Autonomous Government of Andalucia (CTS-7447). M.V.E.-R. is supported by a fellowship (PI11/00533) from ISCIII. We thank all the patients who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figure S1
(JPEG 111 kb)
Supplementary Table S1
(DOC 59 kb)
Supplementary Table S2
(DOC 42 kb)
Supplementary Table S3
(DOC 34 kb)
Supplementary Table S4
(DOC 54 kb)
Rights and permissions
About this article
Cite this article
Torroglosa, A., Enguix-Riego, M., Fernández, R. et al. Involvement of DNMT3B in the pathogenesis of Hirschsprung disease and its possible role as a regulator of neurogenesis in the human enteric nervous system. Genet Med 16, 703–710 (2014). https://doi.org/10.1038/gim.2014.17
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.17
Keywords
This article is cited by
-
Stem cell-based therapy for hirschsprung disease, do we have the guts to treat?
Gene Therapy (2022)
-
Genome-wide analysis of DNA methylation in Hirschsprung enteric precursor cells: unraveling the epigenetic landscape of enteric nervous system development
Clinical Epigenetics (2021)
-
Overexpression of DNMT3b target genes during Enteric Nervous System development contribute to the onset of Hirschsprung disease
Scientific Reports (2017)
-
Identification of different mechanisms leading to PAX6 down-regulation as potential events contributing to the onset of Hirschsprung disease
Scientific Reports (2016)
-
Clinically relevant copy number variations detected in cerebral palsy
Nature Communications (2015)