Abstract
Purpose:
The harmful effects of inbreeding are well known by geneticists, and several studies have already reported cases of intellectual disability caused by recessive variants in consanguineous families. Nevertheless, the effects of inbreeding on the degree of intellectual disability are still poorly investigated. Here, we present a detailed analysis of the homozygosity regions in a cohort of 612 patients with intellectual disabilities of different degrees.
Methods:
We investigated (i) the runs of homozygosity distribution between syndromic and nonsyndromic ID (ii) the effect of runs of homozygosity on the ID degree, using the intelligence quotient score.
Results:
Our data revealed no significant differences in the first analysis; instead we detected significantly larger runs of homozygosity stretches in severe ID compared to nonsevere ID cases (P = 0.007), together with an increase of the percentage of genome covered by runs of homozygosity (P = 0.03).
Conclusion:
In accord with the recent findings regarding autism and other neurological disorders, this study reveals the important role of autosomal recessive variants in intellectual disability. The amount of homozygosity seems to modulate the degree of cognitive impairment despite the intellectual disability cause.
Genet Med 17 5, 396–399.
Similar content being viewed by others
Introduction
Intellectual disability (ID) is one of the most common neuropsychiatric disorders and an unresolved health-care problem, with a prevalence ranging from 1 to 3%.1 The genetic causes of ID have been largely investigated in the past decades, but because of a male prevalence of ID, research was mainly focused on X-linked forms until recently. Molecular karyotyping is the most common technique used today in ID investigation, and gross chromosomal abnormalities explain up to 15% of the cases.2 Despite this, more than 60% of patients have no identifiable genetic cause.3
The recent introduction of next-generation sequencing technologies has opened new possibilities, making the detection of causal variants more economical and faster. Recent studies have revealed that a significant proportion of sporadic ID might be attributable to de novo mutations,4,5 whereas only a small number has shown recessive inheritance. In addition, the vast majority of the studies focused on the role of autosomal recessive variants considered only ID cases in consanguineous families.6,7,8 The widespread availability of high-density single-nucleotide polymorphism (SNP) array data in the past few years has made possible the study runs of homozygosity (ROHs), long genomic stretches of DNA inherited identical-by-descent from the same ancestor that are directly correlated to the inbreeding level.9 In this scenario, ROHs have become a powerful tool for investigating the role of distant inbreeding in any population on a large scale. The cumulative effect of multiple recessive variants, as tested in ROH studies, plays an important role in the etiology of complex diseases. Indeed, excess of ROHs has been identified as a risk factor for Parkinson disease,10 Alzheimer disease,11 schizophrenia,12 and also speech delay and autism.13,14
In this light, we have performed an ROH study on individuals affected by ID with two principal purposes: (i) to test the amount of ROHs as a risk factor for syndromic features of ID and (ii) to investigate the effect of ROHs on the degree of ID.
Materials and Methods
We collected data for 668 (266 female and 402 male) affected children from the pediatric clinics of four Italian hospitals (IRCCS-Burlo Garofolo (TS), IRCCS-Casa Sollievo della Sofferenza (FG), Città della Scienza e della Salute (TO), and IRCCS-Oasi Maria SS (EN)) that are specialized Italian ID health services.
Probands were evaluated with a diagnostic algorithm to exclude cases of nongenetic ID ( Table 1 ) and were then classified as nonsyndromic ID (NSyn-ID), if the only clinical feature detected was the ID, and as syndromic ID (Syn-ID), if additional anomalies were detected. We also collected data regarding neurodevelopmental assessments using the intelligence quotient (IQ) that were evaluated using developmental scales (Bayley III, Brunet-Lezine, or Griffiths) or an intelligence scale (Weschler) according to the age of the patients. We were able to collect IQ information for only 368 subjects, thus excluding the remaining individuals from all analyses using IQ. Individuals were then divided into a nonsevere ID case group (NSev-ID), for IQ ranging from 35 to 75, and a severe ID case group (Sev-ID), for IQ less than 35.
Samples were genotyped using four SNP-array platforms (Illumina San Diego, CA; HumanCytoSNP-12v1, HumanCNV370v1, HumanOmniExpress-12v1, and Affimetrix, Santa Clara, CA; Genome-Wide Human SNP 6.0), depending on the health-care unit providing the samples. Standard genotyping quality control was performed, removing low-quality markers and samples (Supplementary Materials and Methods online).
Given that different ancestral populations have very dissimilar homozygosity distributions,15 we performed principal component analysis and removed outlier individuals (Supplementary Materials and Methods online). After this step, we proceeded with the detection of ROHs using the analysis toolset PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/).16 We searched for continuous stretches of at least 100 homozygous autosomal markers and a minimal length of 1 Mb. We chose 1 Mb as the minimal ROH length because sparser arrays (HumanCytoSNP-12v1 and HumanCNV370v1) are not able to identify shorter stretches. Individuals with ROHs totaling more than 6.25% of the whole genome were excluded because they are typical of descendants of close consanguineous marriages.12 Moreover, to identify possible deletions that would be misclassified as ROHs, we performed copy-number variation detection analysis using PennCNV (http://www.openbioinformatics.org/penncnv/).17
For each subject, we considered (i) the total amount of homozygosity (KB), which is the total Kb spanned by all ROHs, (ii) the mean length of ROH segments (KBAVG), and (iii) the presence of at least one ROH of size >5 Mb.
To overcome differences in ROH distribution attributable to the different array densities, we chose to perform a logistic regression in each data set and then to combine the results in a meta-analysis using a standard inverse variance method in which the contribution of each data set was weighted and correlated with its size.9 The rate of long ROHs in the two groups was calculated regardless of the array type because all the arrays used are able to detect them. The level of significance for the different proportions of long ROHs in the two groups was obtained with the χ2 test.
Results
After a quality-control procedure, 19 individuals were excluded for low-quality data, 37 were removed because of genetic outliers, and 10 were identified as descendants of consanguineous marriages. Moreover, we detected a total of 32 deletions, and the corresponding genomic regions were excluded from further analysis. At the end of these steps, we obtained a clean study sample of 612 individuals. As expected, the distribution of KB and KBAVG differed between the four arrays (Supplementary Table S1 online).
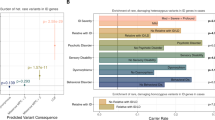
We first investigated the correlation between homozygosity and complexity of phenotype in ID cases. We regressed the syndromic/nonsyndromic status of KB and KBAVG for 612 individuals (187 NSyn-ID and 425 Syn-ID), and no association was detected for KB (odds ratio (OR) = 1.02; confidence interval (CI) = 0.88, 1.20; P = 0.75) or for KBAVG (OR = 0.99; CI = 0.94, 1.04; P = 0.71). The second part of the study focused on the degree of ID (294 NSev-ID and 74 Sev-ID). We found significantly larger homozygous stretches in Sev-ID as compared with NSev-ID cases, together with an increased amount of global homozygosity, despite the lower number of individuals as compared with that of the previous analysis. In Figure 1 , we reported the OR for severe status and the 95% CI from the single logistic regressions and the meta-analysis. Notably, for every 10-Mb increase in total homozygosity, the OR for severe status was increased by 12% (OR = 1.12; CI = 1.01, 1.24; P = 0.030). Moreover, there was a strong association between KBAVG and the degree of ID (OR = 1.08; CI = 1.02, 1.14; P = 0.007). This means that for every 100-kb increase in KBAVG, the OR for Sev-ID status was increased by 8%.
Single data sets and combined analysis for global homozygosity and mean length of ROH stretches. For each analysis, we reported the number of individuals (n), the estimated changes in odds ratio (OR), and the 95% confidence intervals (95% CIs). On right side, we reported the meta-analysis forest plots. Squares represent the OR for each data set, and their size is proportional to contribution of each data set; horizontal lines are the respective CIs. The diamond represents the summary measurement. In a, the result of global homozygosity analysis is reported, and an increase of 10 Mb in KB was considered. In b, the mean length of ROH is reported, with the increase in KBAVG set to 100 Kb. In both cases, the summary measurements were significantly >1, although it is clear that the signal is stronger in b than in a. KBAVG, the mean length of ROH segments; OR, odds ratio; ROH, run of homozygosity.
To check for the presence of confounding factors, several models were tested that included sex, the first three principal components for possible population stratification, sample origin, and Syn-ID/NSyn-ID status; statistical significance for KBAVG was obtained for all the tests. The effect of larger ROHs was confirmed by counting the number of subjects with at least one ROH longer than 5 Mb; the rates of those subjects were 0.24 for Sev-ID patients and 0.12 for NSev-ID patients (P = 0.0087). Such long ROHs accounted for 20% of total homozygosity across Sev-ID individuals and for only 6% in NSev-ID individuals. Finally, we checked for ROHs frequently present in Sev-ID as compared with NSev-ID, but we did not find any significant association (Supplementary Table S2 online).
Discussion
The harmful effects of inbreeding are well known by geneticists. Sons of consanguineous couples have an increased probability of being affected by a recessive pathology attributable to identical-by-descent mutations, and this predisposition is inversely correlated with the distance from the common ancestor. Although it was first noted in fitness-related phenotypes, several recent studies identified the amount and the size of ROHs, including homozygosity arising from distant inbreeding, as an important player in neurological disorders. Nevertheless, the way in which ROHs contribute to the etiology of the disease is still unclear.
Following this approach, we tested the effect of ROHs on the presence of syndromic features in individuals with ID who were not descendants of consanguineous mating. Syndromic conditions are more common in very close inbreeding and, although one mutation can be the cause of Syn-ID, different features may be attributable to multiple recessive homozygous mutations in different genes. Based on our analysis, ROHs are not associated with the complexity of the phenotype (e.g., malformations, seizure, and microcephaly/macrocephaly). A possible explanation is that in our cohorts the majority of syndromic cases were not the result of multiple gene defects but instead were attributable to single-gene ID syndromes. However, we detected significant associations between both total quantity and mean length of ROHs with the degree of ID, which is not biased by the presence of cases of extremely close inbreeding because, as a precaution, these were removed from our data set. This correlation between long ROHs and severity of ID is even stronger when considering only ROHs larger than 5 Mb. Not surprisingly, our results are very similar to the effects of homozygosity on ID in simplex autism.13
The stronger effect of length with respect to total amount of ROHs can be explained with two mechanisms. On the one hand, we had limited data and we could not exclude an overestimation of the extent of the ROHs because of the lack of array resolution and, for the same reason, we could not analyze ROHs shorter than 1 Mb. Because we were missing shorter stretches, the associations were driven by long ROHs because they are easily detected. On the other hand, it is possible that longer ROHs, which arise from closer inbreeding, could have a greater effect, leaving an open question regarding the influence of short ROHs on ID. According to Szpiech et al.,18 who reported an enrichment of deleterious variants in long ROHs in healthy individuals, long stretches of homozygosity may contain more likely damaging variants arising from close inbreeding that have not yet been purged by selection. Those variants may have an important role in ID modulation.
Further investigation would require testing our findings in a larger cohort using whole-genome sequencing analysis. In this way it would be possible to obtain a more accurate estimate of the extent of ROHs, especially for short stretches. In addition, our results indicate that homozygosity mapping on sequencing data can be a valuable tool to identify genes involved in modulation of the degree of ID. The most important consequence of this study is probably that, despite the heterogeneity of ID causes in our cohorts (copy-number variation, insertions/deletions, and single-nucleotide variations), the distribution of homozygosity seems to have a significant impact on determining the severity of impairment, implying an important contribution of recessive genetics effects. This is not surprising because cognitive ability is a very complex trait and its total variability is unlikely to be determined by a single pathogenic mutation. Other environmental and genetic factors could modulate the phenotype, and one of the most important seems to be accounted for by ROHs.
Disclosure
The authors declare no conflict of interest.
References
Leonard H, Wen X . The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 2002;8:117–134.
Hochstenbach R, Buizer-Voskamp JE, Vorstman JA, Ophoff RA . Genome arrays for the detection of copy number variations in idiopathic mental retardation, idiopathic generalized epilepsy and neuropsychiatric disorders: lessons for diagnostic workflow and research. Cytogenet Genome Res 2011;135:174–202.
Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 2006;140:2063–2074.
Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet 2010;42:1109–1112.
de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012;367:1921–1929.
Abou Jamra R, Wohlfart S, Zweier M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet 2011;19:1161–1166.
Çalışkan M, Chong JX, Uricchio L, et al. Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Hum Mol Genet 2011;20:1285–1289.
Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011;478:57–63.
McQuillan R, Eklund N, Pirastu N, et al.; ROHgen Consortium. Evidence of inbreeding depression on human height. PLoS Genet 2012;8:e1002655.
Simón-Sánchez J, Kilarski LL, Nalls MA, et al.; International Parkinson’s Disease Genomics Consortium; Wellcome Trust Case Control Consortium. Cooperative genome-wide analysis shows increased homozygosity in early onset Parkinson’s disease. PLoS One 2012;7:e28787.
Ghani M, Sato C, Lee JH, et al. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol 2013;70:1261–1267.
Keller MC, Simonson MA, Ripke S, et al.; Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet 2012;8:e1002656.
Lin PI, Kuo PH, Chen CH, et al. Runs of homozygosity associated with speech delay in autism in a Taiwanese Han population: evidence for the recessive model. PLoS One 2013;8:e72056.
Gamsiz ED, Viscidi EW, Frederick AM, et al.; Simons Simplex Collection Genetics Consortium. Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am J Hum Genet 2013;93:103–109.
Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF . Genomic runs of homozygosity record population history and consanguinity. PLoS One 2010;5:e13996.
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575.
Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–1674.
Szpiech ZA, Xu J, Pemberton TJ, et al. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet 2013;93:90–102.
Acknowledgements
The authors thank Stefania Cappellani, Lisa Cleva, and Anna Morgan. This study was made possible by the financial support of the Research Project n.33/2011 “identification of new genes involved in intellectual disability.”
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
(DOC 53 kb)
Rights and permissions
About this article
Cite this article
Gandin, I., Faletra, F., Faletra, F. et al. Excess of runs of homozygosity is associated with severe cognitive impairment in intellectual disability. Genet Med 17, 396–399 (2015). https://doi.org/10.1038/gim.2014.118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.118
Keywords
This article is cited by
-
Long contiguous stretches of homozygosity detected by chromosomal microarrays (CMA) in patients with neurodevelopmental disorders in the South of Brazil
BMC Medical Genomics (2019)
-
Genome-wide tracts of homozygosity and exome analyses reveal repetitive elements with Barrets esophagus/esophageal adenocarcinoma risk
BMC Bioinformatics (2019)
-
Runs of homozygosity: windows into population history and trait architecture
Nature Reviews Genetics (2018)
-
Whole genome sequencing identifies missense mutation in MTBP in Shar-Pei affected with Autoinflammatory Disease (SPAID)
BMC Genomics (2017)
-
Long contiguous stretches of homozygosity spanning shortly the imprinted loci are associated with intellectual disability, autism and/or epilepsy
Molecular Cytogenetics (2015)