Abstract
The Evaluation of Genomic Applications in Practice and Prevention Working Group was first convened in 2005 to develop and test evidence-based methods for the evaluation of genomic tests in transition from research to clinical and public health practice. Over the ensuing years, the Working Group has met 26 times, publishing eight recommendation statements, two methods papers, and one outcomes paper, as well as planning and serving as technical experts on numerous associated systematic reviews. Evaluation of Genomic Applications in Practice and Prevention methods have evolved to address implications of the proliferation of genome-wide association studies and are currently expanding to face challenges expected from clinical implementation of whole-genome sequencing tests. In this article, we review the work of the Evaluation of Genomic Applications in Practice and Prevention Working Group over the first 8 years of its existence with an emphasis on lessons learned throughout the process. It is hoped that in addition to the published methods of the Working Group, the lessons we have learned along the way will be informative to others who are producers and consumers of evidence-based guidelines in the field of genomic medicine.
Genet Med 2014:16(3):217–224.
Similar content being viewed by others
Main
The completion of the human genome project was heralded as the dawn of the era of genomic-based personalized medicine. Numerous factors, however, have complicated the translation of scientific findings into clinical genomic testing with measurable health outcomes. The responsible integration of genomic technologies into medical care poses challenges to health-care providers, consumers, and other stakeholders. These emerging genomic applications to health care have been discovered by a scientific community that may have complex ties to industry and that is championed by strong public advocates and protected from rigorous scrutiny under the philosophy of genetic exceptionalism; in addition, these genomic applications are provided via direct access through advertising to a public with few resources for objective information.
Nonetheless, the potential of the human genome project to fundamentally change our understanding of disease causation cannot be overstated. The ability to tailor clinical and public health interventions to individuals or populations on the basis of their predisposition to diseases or response to treatment remains an inspiring goal of genomic medicine, and novel genomic diagnostics are allowing molecular targeting of therapies. As in any translation of new technology to health care, critical issues in genomic testing are now being defined. Little consensus exists among key stakeholders regarding the framework for developing, implementing, and evaluating genomic testing, and there are often sparse clinical data supporting the utilization of genomic testing in caring for patients. There are gaps in knowledge and oversight that could lead to serious harms.1 While working toward the realization of the potential of genomics to improve health, it is imperative to implement processes that can protect individuals from the potential harms of premature implementation of genomic testing while supporting innovations that may produce significant improvements in health. It was in response to this imperative that the Centers for Disease Control and Prevention created the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative. The EGAPP initiative was designed to support the translation of scientific evidence on genomic testing into clinical practice. This article outlines the lessons learned from the EGAPP initiative, in the context of efforts of various professional, governmental, and private entities to consider how to advance genomic science along the translational continuum from bench to bedside while also addressing current and future challenges to genomics research.
The Office of Public Health Genomics and the EGAPP Initiative
The Centers for Disease Control and Prevention established the Office of Public Health Genomics (OPHG) in 1997 to address how advances in genomics could most appropriately be used across populations to improve health and prevent disease. This program was designed to support the translation of findings from genomic studies to meet population-level health needs.
The OPHG sponsored the ACCE project (Analytic validity, Clinical validity, Clinical utility, and Ethical, legal, and social implications) in 2000, which created a model for evaluating the use of genetic tests, applicable across multiple tests and testing scenarios. The ACCE project involves evaluating the test and its application to a specific clinical scenario by examining analytic validity (how well does the test measure what it is supposed to measure?), clinical validity (how well does the test predict the phenotype of concern, e.g., risk of disease, prognosis, or response to therapy?), and clinical utility (does clinical use of the test provide a net health benefit?) while considering ethical, legal, and social implications of using the test in individual patients.2
Building on the results of the ACCE project, the OPHG established the EGAPP initiative in 2004 to create a methodology for evaluating the evidence on genomic tests. The EGAPP Working Group (EWG) was formed as an independent panel charged with developing and testing systematic processes for the evidence-based evaluation of genomic tests and the translation of these evaluations into evidence-based recommendations for clinical use. The ACCE project created the framework for answering questions about genetic tests. By contrast, the EWG was established to develop the methods for evidence grading for analytic validity, clinical validity, and clinical utility for generating clinical recommendations for the use of genomic tests in multiple clinical and public health scenarios.
The EWG was originally intended to design and test methods that would be applicable to the evaluation of the rapidly expanding number of genomic tests available for common diseases. It was not intended to be an ongoing program to evaluate and synthesize evidence or to create and maintain a comprehensive set of clinical recommendations as the US Preventive Services Task Force does for other clinical preventive services. The EWG’s founding members included experts in evidence-based review, clinical practice, clinical guideline development, public health, laboratory practice, genomics, epidemiology, economics, ethics, policy, and health technology assessment. EWG work was organized around methods development and testing, horizon scanning for new tests and topic prioritization, and development and dissemination of findings, including clinical recommendations based on systematic evidence syntheses.
EGAPP Lessons Learned: Methods
The EWG 2009 methods paper3 detailed the challenges for evidence-based approaches to genomics: many disorders targeted by genomic tests are uncommon; technologies rapidly emerge and evolve; clinical applications vary substantially (e.g., for presymptomatic risk assessment, diagnosis, screening, prognosis, or pharmacogenomics); outcomes are often ill defined; interpreting test results is often complex; and genomic test results may have implications for multiple family members. For example, the clinical and family implications may be clear for some tests, such as those for Lynch syndrome to guide colorectal cancer screening services for family members. However, clinical implications are more complex for untreatable diseases such as Huntington disease, for which the primary outcomes of testing may involve reproductive choices or planning for the burden of a future, inevitable disease.
It was apparent from the EWG’s methods work that the relevant stakeholders in genomics had disparate professional interests, languages, and norms. Various professional groups, for example, had fundamental differences in how they valued data. Although many geneticists and laboratory scientists believed that the information from testing had value in and of itself, those tied to an evidence-based perspective believed that data are only valuable if they can be linked to important and measurable health outcomes. Having made a decision to focus on a test’s ability to influence important health outcomes, the EWG also faced key issues including how, whether, and when to use contextual information, such as the prevalence and severity of the disease under consideration; the cost and feasibility of testing and treatment; and the implications of not testing. Although strict evidence-based recommendations may be based solely on effectiveness or efficacy, contextual information can be an important aspect of the decision-making process.
Availability and applicability of evidence
Implementing the new EWG methods underscored the limits of available genomic research in core areas of the analytic framework used in EGAPP reviews. Systematic review of analytic validity research, for example, highlighted the need to try to obtain, evaluate, and summarize unpublished information about laboratory methods, test reliability, sample rejection, proficiency testing, and other data that were held by the developing laboratories and their related proprietary interests. Traditional systematic evidence review methods were insufficiently robust to guide effective searching of the unpublished “gray literature,” nor did they provide an adequate means for rating the quality of these data or summarizing them, because the typical synthesis techniques used for clinical validity and utility studies could not be applied. In the case of tumor gene expression profiling in breast cancer, there were some published data on analytic validity; however, there was no “gold standard” for comparison that would permit estimation of analytic sensitivity or specificity.4 For many of the recommendation statements, the EWG concluded that the evidence to assess analytic validity was insufficient. As a result, the EWG often developed recommendations on the basis of more complete evidence further along the analytic framework (i.e., clinical validity and clinical utility). The EWG recognized inherent risks in assuming adequate levels of analytic validity, and the possibility that clinical trials and clinical trial results might be based on erroneous assumptions regarding analytical validity. The unfortunate experience with Duke University and the harms associated with undertaking clinical trials on the basis of flawed research underscore the critical nature of ensuring analytic validity.5
Moreover, data on clinical validity were often problematic. For example, in evaluating the effectiveness of cytochrome P450 (CYP450) testing to predict response to selective serotonin reuptake inhibitor therapy, the association of drug metabolism with certain mutations was assumed to translate to management utility. However, the evidence of an association between CYP450 variants indicating metabolizer status and blood levels of selective serotonin reuptake inhibitors was poor, and in fact, no relationship between metabolizer status and selective serotonin reuptake inhibitor treatment outcomes had been reported. Therefore, one could have concluded that the evidence of clinical validity was insufficient to even support clinical trials of utility.6
Applying traditional evidence-based medicine methods to assess clinical utility presented additional challenges. Defining the clinical scenario is particularly important and can be complex. When evaluating the use of UGT1A1 testing to guide irinotecan therapy for colon cancer, the EWG found adequate evidence for analytic validity, and adequate although mixed evidence for clinical validity. The clinical scenario put forth, however, was to use test results to lower irinotecan doses for slow metabolizers to decrease the risk of adverse effects from elevated drug levels. However, it is possible that a more compelling case could be made for using test results to increase the doses for fast metabolizers in order to increase remission rates.7 This balance between efficacy and adverse events will be an important recurring issue, providing the potential for harm through the promotion of less-effective therapies.
The most common problem with assessing clinical utility, however, was the paucity of good-quality evidence with which to generate an evidence-based recommendation. This is also a recurring theme and remains a critical issue underlying the successful translation of genomics into improved individual and population health. Our gold standard of evidence, the randomized controlled trial, is constrained by time and resource requirements and can be challenging in the face of rare conditions and relatively small effect sizes as compared with usual care. Existing EWG methods do allow for a determination of adequate clinical utility based on a single nonrandomized controlled trial or even a systematic review based on cohort studies alone, when results warrant.3 The EWG experience supports the need for additional approaches and methods for evidence generation, such as consideration of novel observational study designs and innovative modeling strategies, along with a careful examination of the evidentiary standards for clinical recommendations. However, basing recommendations on evidence from studies with lower levels of internal validity will, by definition, require accepting a higher risk of being wrong, providing no net health benefit or even doing more harm than good. Such recommendations will need to be made with a careful consideration of the risk and potential magnitude of harms balanced with the opportunity and potential magnitude of benefits, a plan for addressing the outstanding evidence gaps with additional research, and the discipline to change the guideline and clinical practice if future studies find us wrong.
Improving efficiency
Given the resource requirements of a traditional systematic review, and the sparse evidence on many genomic tests, the EWG also sought to develop and test more efficient and less resource-intensive methods in an attempt to produce shorter, more timely, and less costly targeted or brief reviews on nominated topics. Although brief reviews were intended to be as thorough as full reviews, they offered the potential advantages of faster completion to meet the needs of an area of rapidly evolving research and clinical practice. These brief reviews were intended to be more focused in scope and were to be used when available research was not robust or when a decision regarding the research question or clinical scenario involved only a single aspect of the EWG methods, such as clinical validity. The intent was to produce recommendations that would provide users with the essential information on an application and point out important gaps in research. In practice, however, each targeted review had to be comprehensive in order to support a reliable recommendation, and the EWG discovered that they were still time and resource intensive. It is clear that methods still need to be developed to address issues of timeliness and adequate resources for creating evidence-based guidance for genomic testing.
Modeling
The use of modeling represents an emerging tool in evidence-based medicine methodology and was adopted as a strategy that the EWG could employ in certain circumstances when evaluating genomic applications. Because many of the test applications address uncommon conditions, for which testing and the subsequent cascade of diagnosis and treatment events may cause harms, modeling provides a quantitative, clinically meaningful estimate of the risk trade-offs. False-positive tests, for example, may lead to unnecessary and potentially harmful diagnostic procedures or treatments. In the setting of rare conditions, false positives significantly outnumber true positives, potentially having a great impact on the balance of the benefits and harms of testing. Modeling also enables the EWG to incorporate research-based estimates of real-world test performance (including sensitivity, specificity, and predictive values) along with downstream benefits and harms.8 Although only basic modeling techniques have been used by EWG to date, there is interest among the group in further exploring the performance of more sophisticated modeling schemes in genomic test evaluation. A collaborative effort between the EWG, National Cancer Institute’s Cancer Intervention and Surveillance Modeling Network, and investigators at Kaiser Permanente Center for Health Research aimed at modeling the potential added value of single-nucleotide polymorphisms (SNPs) and family history in colorectal cancer screening is currently under way.
Association studies
The rapid proliferation of genome-wide association studies (GWASs), which has occurred since the inception of the EGAPP, has also provided the EWG with unique methods challenges. Many SNPs are found to have statistically significant associations with common chronic diseases such as cardiovascular disease and type 2 diabetes. The vast majority of the SNPs evaluated by the EWG have relative risks for disease in the 1.1–1.3 range. GWASs are not designed to address the biologic mechanism through which these variants translate to increased risk and, for the most part, there were no high-quality studies to suggest that differential management of individuals with these mutations translates to improved health outcomes. Indeed, where studies had been performed, first-generation GWAS-derived common variants did not add significantly to receiver-operating characteristic curves for common diseases such as breast cancer. At the same time, GWASs had led to the marketing of risk-profiling applications offered through direct-to-consumer marketing strategies. The methods developed for analyzing these GWAS-based risk-profiling approaches8 demonstrated that, given the low combined predictive value of small-risk SNPs, it would be extremely unlikely that any profile would translate to a meaningful increase in overall individual risk for disease. Therefore, management decisions based on these profiles could not be expected to translate to improvements in health outcomes. Moreover, there were important but unanswered questions about whether such testing added value to traditional risk factor assessment (e.g., family history, lipid levels) and whether studies of clinical utility were able to demonstrate that the treatment of individuals who might be reclassified as potentially benefitting from therapy actually experienced improved health outcomes. For example, cardiovascular disease risk is traditionally assessed through the Framingham risk assessment of nongenetic risk factors. Cardiogenomic risk profiling continues to be offered for risk assessment. Although there are some clinical validity studies that suggest potential reclassification from low or intermediate Framingham risk to high risk (based mainly on 9p21 variants), there are no studies that demonstrate that more aggressive cholesterol therapy in individuals so reclassified actually improves heart disease outcomes.9 An alternative use of risk stratification would be to identify individuals at a lower relative risk of a condition that could potentially lead to a recommendation not to undergo population-based screening for a condition that, in theory, could lower costs while not affecting outcomes (e.g., recommending against colonoscopy due to lower risk of colorectal cancer or delaying initiation of mammography due to lower risk of breast cancer). Preliminary reviews have not identified compelling evidence that this outcome is likely, although this question will be examined further in the future. As with other uses of genomic risk stratification, such approaches would still require well-designed trials able to directly assess the benefits and hazards of such interventions. Although GWAS remains an efficient method for describing disease–SNP relationships, translating the results of these studies into polygenic risk assessments that improve health will require more research, a better understanding of the underlying disease mechanisms, and profiles that define more significant levels of increased risk.
Quantifying the harms of withholding therapy
Finally, the EWG learned important methodology lessons for the evaluation of gene expression profiling as a basis for therapeutic decision making. When evaluating the use of breast cancer tumor gene expression profiling for risk stratification following surgery, the systematic evidence review10 found sufficient evidence for clinical validity but found areas of concern in terms of clinical utility. In this setting, profiles were being used to identify women who were unlikely to benefit from postoperative adjuvant chemotherapy in order to avoid unnecessarily exposing them to the risk of adverse effects. The EWG was concerned that a lower likelihood of benefit did not translate to zero chance of benefit and that there was a small but real risk that, with the use of the tests, some women would be denied life-extending therapy. The EWG process could not balance this potential harm against the benefits of avoided side effects. Therefore, the EWG concluded that the evidence was insufficient to recommend routine use of this already commonly used strategy for guiding therapy.4
EGAPP Lessons Learned: Outcomes
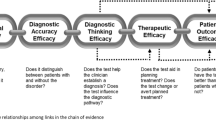
The EWG also realized that our work required detailed definitions of the outcomes that should be considered in assessing the use of genomic tests. The EWG discussed these outcomes in a paper, articulating that outcomes should inform recommendations that balance potential health benefits and harms but that they also must be considered in relation to their relative importance to patients and providers.11 Outcomes were organized into four groups that represent the logical flow from patient impact to broad societal impact: (i) diagnostic thinking/health information impact, (ii) therapeutic choice, (iii) patient outcome impact, and (iv) familial and societal impact. These considerations ultimately provided the EWG with tools used to better consider the indications for use of a test, the surrounding clinical context, and the value of the information to be gained. Although a formal checklist for rating potential outcomes, arranged according to the four domains mentioned above, was created, we found that the thought process involved in developing the list was at least as helpful as completing it. We have found that informal approaches to consideration of outcomes, contextual issues, and ethical, legal, and social implications, as long as they are discussed throughout the evaluation process, can add value to rigorous evaluation methods.
EGAPP Lessons Learned: Horizon Scanning
Due to the rapid development of scientific evidence for genomic tests and the lack of a centralized repository for genomics information, the EWG recognized that methods for actively monitoring changes in this evidence are extremely important to producing timely evaluations. Initially the process for evidence scanning was informally organized and primarily involved panel members gathering information through professional interactions, such as meetings, conferences, and abstracts. In 2009, OPHG staff assumed responsibility for developing and implementing a more formal process for horizon scanning. Searches now rely on the GAPP Knowledge Base, an openly accessible online database of available genomic tests that also includes gray literature searches of Internet sources, newspapers, and blogs.12
EGAPP Lessons Learned: Stakeholder Engagement
The EGAPP Stakeholders Group (ESG) was created to enable the EWG to work directly with stakeholders in formulating review questions, helping to guide evidence reviews with pertinent information, and informing recommendation development. The ESG included not only representatives from similar areas as the EWG but also professional society and industry representatives and advocates who had previously worked with the EGAPP, including patient advocacy groups. As the relationship between the two groups developed, the EWG felt that the ESG expected a level of input into the conduct of the review and the recommendation that was inconsistent with maintaining a strict, evidence-based process. For example, the ESG provided feedback to the EWG that the format of the recommendation statements was not effective in communicating the message to stakeholders, but this was perceived by the EWG as altering the evidence-based recommendations. Unfortunately, a confluence of suboptimal communication and other factors resulted in the disbanding of the ESG. Retrospectively, a clearer description of the roles, responsibilities, and management of expectations of the participants at the outset, as well as a more coordinated creation of both groups, may have resulted in an important and valuable working relationship. Stakeholder involvement and increased transparency is critical to the successful translation of evidence-based recommendations into acceptable medical practice, and new, major research efforts such as the Patient-Centered Outcomes Research Institute are wrestling with this new imperative. The EGAPP experience underscores how difficult this work can and will be.
On the basis of these lessons from the ESG, and on the continued desire for outside expert input, the Centers for Disease Control and Prevention and National Institutes of Health created the Genomic Applications in Practice and Prevention Network. This network was designed as an open-member, collaborative network that would build the evidence base for genomic technologies and also help translate study results into practice. This network provided an avenue through which the EWG could communicate credible information to and solicit feedback from stakeholders. The network is intended to “accelerate and streamline effective and responsible use of validated and useful genomic knowledge and applications, such as genomic tests, technologies, and family history, into clinical and public health practice”13 and therefore complements the EWG’s work and research agenda. The near future of the network is uncertain because resource constraints are likely to influence the full realization of this network.
EGAPP Lessons Learned: Clinical Recommendations
The dissemination of recommendations and their impact on the knowledge of health professionals and the public are essential aspects of the EGAPP initiative’s mission. The first eight completed recommendation statements are summarized in Table 1 , which lists the clinical scenario or setting for testing, the finding and recommendation, and for those topics for which insufficient evidence was found to recommend for or against testing, a clinical context statement regarding current use or further study. The EGAPP initiative has tracked the potential for impact of selected recommendations through media and literature searches. These revealed that EGAPP recommendations have generated hundreds of articles in professional and consumer publications ranging from online press releases to editorials in peer-reviewed journals. The CYP450 recommendation, for example, was cited in more than 75 articles in a broad range of publications, thus demonstrating the potential for this information to influence practice and policy. The ensuing articles clearly explicated that the evidence did not support CYP450 screening to aid in antidepressant selection. Several of these articles also articulated some of the difficulties the EGAPP initiative faces when evaluating genomic innovation. In doing so, they not only helped dissemination of EGAPP recommendations but also started a public conversation on evidence-based methodology for genomic applications. The recommendations also caught the attention of researchers and clinicians, many of whom were critical of both EGAPP methods and results. This supported the academic dialogue necessary to refine and improve evidence-based methods and to champion their application in genomic medicine.
EGAPP Lessons Learned: Evidence Practice Centers
The EWG worked with Agency for Healthcare Research and Quality Evidence-Based Practice Centers (EPCs) to conduct systematic evidence reviews of topics of interest. Trying to apply the language of genomics to the existing framework of the Agency for Healthcare Research and Quality evidence centers was challenging, and some of the standard EPC methods were not ideal for this area. For example, the analytic validity of tests used in general medical practice is often assumed in EPC reviews without in-depth review but is a critical issue for genomics, in which technology is rapidly changing and interpretation of results is critical but not necessarily well standardized. EPC methods are time consuming, making it difficult for those applying them to keep up with the speed of genomic discovery and innovation; reviews and evidence-based recommendations can easily be out of date by the time they are published. The use of a single genomic test can evolve so quickly that research data are often very heterogeneous, requiring the expertise of different disciplines to help interpret the information. This type of specific genomics and clinical expertise was not always readily available at the more generalist EPCs.
Learning from the EPC experiences, the Centers for Disease Control and Prevention funded an EGAPP-specific EPC-like entity, named the Knowledge Synthesis Center. Requiring innovative methodology and organization to face the EGAPP initiative’s unique challenges, the Knowledge Synthesis Center is intended to work directly with the EWG much like the single EPC that supports the majority of the US Preventive Services Task Force’s work, allowing for efficient communication, clear expectations, and appropriately tailored expertise, all required elements when applying evidence-based methods to a rapidly evolving body of research. The Knowledge Synthesis Center approach has been very successful to date, working with the EWG on several new projects that have been submitted for review or are in press.
EGAPP Lessons Learned: Membership, Direction, and Scope
By the end of the third year of the EGAPP initiative, the content of the work suggested the need for additional experts from a broader range of disciplines. Given the technical nature of genomics testing and the degree to which it was being used in cancer risk assessment, screening, and treatment, new members from the disciplines of oncology and molecular pathology were recruited. A better understanding of the complexity of pharmacogenomics led to the addition of a national expert. It remains a constant challenge to identify key specialty areas where new or additional representation is needed on the EWG and to discern those instances for which a need for expertise can be best met through strategic collaborations rather than panel membership.
The EWG has published eight evidence-based recommendation statements, two methods papers, and one outcomes paper since its inception, largely succeeding in the initial goal of developing and testing evidence-based medicine methods for use in genomic testing. In 2008, the EWG revisited its basic direction for continuing work, considering whether the primary role of the EWG should remain development and evaluation of evidence-based methods or whether it should shift toward continuous and ongoing recommendation development. EWG members concluded that assuming the role of an active recommendations panel, such as the US Preventive Services Task Force, would require a significant increase in resources, and it was unclear if stakeholders were interested in this level of commitment and investment; this issue remains unresolved. The EWG remains committed and enthusiastic about contributing toward moving the field forward, and at least in the near term, this may be best accomplished through a multifunctional approach.
The EGAPP Initiative Today
The integration of genomics into medicine continues to need an objective and independent process and resource, such as the EGAPP initiative, to evaluate evidence and provide stakeholders with evidence-based recommendations. However, economic conditions brought both critical federal budget concerns and a need for additional focus in public health programs. The funding for OPHG and the EGAPP programs was significantly reduced. The implementation of next-generation sequencing techniques will provide more affordable and available approaches to large-scale genome sequencing, presenting clinicians and patients with a vast amount of new information of unknown and unproven health value, with continued significant potential for health harms and increased costs. The need for an evidence-based “honest broker” for reviewing and synthesizing evidence and translating it into clinical recommendations remains as important as ever. Using an evidence-based, systematic approach to understanding how such information can and should be used is imperative.
The members of the EWG remain committed to advancing the agenda of evidence-based genomic testing and developing relationships with new partners to ensure that our work informs the work of those that follow. The EWG continues to develop systematic review-based recommendation statements concerning the clinical use of genomic tests. Specific projects could involve applying EGAPP methods in areas such as categorizing elements of the genome into bins that relate to clinical use14 and addressing specific screening topics for which there could be significant family history and genetic influences, such as colorectal cancer screening. Initial collaborative work on the former has already begun between the EWG and the Knowledge Synthesis Center, and an initial article describing development and pilot testing of a method for reporting incidental findings from whole-genome sequencing tests has been published.15 Regardless of how the EGAPP initiative evolves, the lessons learned from it should be embraced by those leaders envisioning, developing, and implementing the discipline of genomic medicine to ensure that we fully realize the promise and opportunity for better health inspired by the decoding of the human genome.
Summary of Lessons Learned from the EGAPP Initiative
The points are given as follows:
-
1
Evidence on analytic validity is sparse, often found only in the gray literature or completely unavailable due to proprietary concerns, and not easily assessed with traditional evidence synthesis methods.
-
2
Evidence on clinical utility needs to focus on associations with clinically important intermediate or distal health outcomes.
-
3
Evidence on clinical validity of genomic tests is sparse and not of the quality usually found in support of other evidence-based clinical recommendations.
-
4
In responding to the rapidly evolving generation of new genomic testing scenarios and supporting evidence, it is challenging to employ the traditional evidence review and synthesis methods with less time and limited resources.
-
5
Modeling approaches can provide important quantitative estimates of benefits and harms that can inform recommendations for genomic testing.
-
6
Genetic risk profiling based on GWAS study results is unlikely, as currently developed and implemented, to translate into management decisions that improve individual health or provide significant additional information as compared with traditional risk assessment.
-
7
Benefits of avoiding less aggressive and less costly therapy based on genomic testing must adequately quantify the individual risks of harm associated with withholding that therapy.
-
8
In assessing genomic testing strategies, it is essential to clearly outline the relevant outcomes to consider, such as (i) diagnostic thinking/health information impact, (ii) therapeutic choice, (iii) patient outcome impact, and (iv) familial and societal impact.
-
9
Monitoring the evolving horizon of new genomic tests and supporting evidence is essential to guiding ongoing evaluation of genomic medicine approaches.
-
10
Stakeholder involvement in creating recommendations for genomic testing is important and difficult, and a clear description of roles, responsibilities, and expectations may better support success.
-
11
EPCs need specific expertise to support evidence review and synthesis as well as recommendation development for genomic testing.
-
12
Membership of panels that engage in evidence-based recommendation development must be representative of all the ethical, clinical, and other scientific disciplines related to the specific area of genomic testing.
Disclosure
D.L.V. reports that he was a consultant for Medco, Novartis Molecular Diagnostics, and Genentech and is supported by the following genomics-related research grants: P50HG003374, RC2CA148570, UO1GM092676, and UO1HG006507 from the National Institutes of Health and U18GD000005 from the Centers for Disease Control and Prevention. S.G.P. reports that one of his research studies was supported by a fund from Novartis to Tufts Medical Center. S.R.T. has no personal conflicts of interest to disclose. The Center for Medical Technology Policy receives funding from several sources, listed at http://www.cmtpnet.org/about/funding-sources/. The other authors declare no conflict of interest.
References
Committee on Developing Biomarker-Based Tools for Cancer Screening, Diagnosis, and Treatment. Nass SJ, Moses HL (eds). Cancer Biomarkers: The Promises and Challenges of Improving Detection and Treatment. National Academies Press: Washington, DC, 2007.
Haddow J, Palomaki G . ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury M, Little J, Burke W (eds). Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. Oxford University Press: New York, 2003:217–233.
Teutsch SM, Bradley LA, Palomaki GE, et al.; EGAPP Working Group. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med 2009;11:3–14.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet Med 2009;11:66–73.
Baggerly KA, Coombes KR . What information should be required to support clinical “omics” publications? Clin Chem 2011;57:688–690.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: testing for cytochrome P450 (CYP450) polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med 2007;9:819–825.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? Genet Med 2009;11:15–20.
Veenstra DL, Piper M, Haddow JE, et al. Improving the efficiency and relevance of evidence-based recommendations in the era of whole-genome sequencing: an EGAPP methods update. Genet Med 2013;15:14–24.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: genomic profiling to assess cardiovascular risk to improve cardiovascular health. Evid Rep Technol Assess 2007;12:839–843.
Marchionni L, Wilson RF, Marinopoulos SS, et al. Impact of gene expression profiling tests on breast cancer outcomes. Evid Rep Technol Assess 2007;160:1–105.
Botkin JR, Teutsch SM, Kaye CI, et al.; EGAPP Working Group. Outcomes of interest in evidence-based evaluations of genetic tests. Genet Med 2010;12:228–235.
GAPP Finder website. http://www.hugenavigator.net/GAPPKB/topicStartPage.do Accessed 21 May 2013.
GAPPNet website. http://www.gappnet.org/ Accessed 20 December 2012.
Berg JS, Khoury MJ, Evans JP . Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med 2011;13:499–504.
Goddard KA, Whitlock EP, Berg JS, et al. Description and pilot results from a novel method for evaluating return of incidental findings from next-generation sequencing technologies. Genet Med 2013; e-pub ahead of print 4 April 2013.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: does genomic profiling to assess type 2 diabetes risk improve health outcomes? Genet Med 2013; 15: 612–617.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy? Genet Med 2013;15:517–527.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: routine testing for factor V Leiden (R506Q) and prothrombin (20210G>A) mutations in adults with a history of idiopathic venous thromboembolism and their adult family members. Genet Med 2011;13:67–76.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 2009;11:35–41.
Acknowledgements
The authors thank Kevin Lutz, Kaiser Permanente Research Affiliates Evidence-Based Practice Center and Knowledge Synthesis Center, Center for Health Research, Kaiser Permanente Northwest, for valuable assistance in the preparation of this article.
Author information
Authors and Affiliations
Consortia
Rights and permissions
About this article
Cite this article
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. The EGAPP initiative: lessons learned. Genet Med 16, 217–224 (2014). https://doi.org/10.1038/gim.2013.110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2013.110
Keywords
This article is cited by
-
Measuring clinical utility in the context of genetic testing: a scoping review
European Journal of Human Genetics (2021)
-
The Routine Clinical use of Pharmacogenetic Tests: What it Will Require?
Pharmaceutical Research (2017)
-
Pharmacogenetic approaches in the treatment of alcohol use disorders: addressing clinical utility and implementation thresholds
Addiction Science & Clinical Practice (2014)