Abstract
Purpose:
The aim of the study was to evaluate the diagnostic accuracy of an informatics-based, noninvasive, prenatal paternity test using array-based single-nucleotide polymorphism measurements of cell-free DNA isolated from maternal plasma.
Methods:
Blood samples were taken from 21 adult pregnant women (with gestational ages between 6 and 21 weeks), and a genetic sample was taken from the corresponding biological fathers. Paternity was confirmed by genetic testing of the infant, products of conception, control of fertilization, and/or preimplantation genetic diagnosis during in vitro fertilization. Parental DNA samples and maternal plasma cell-free DNA were amplified and analyzed using a HumanCytoSNP-12 array. An informatics-based method measured single-nucleotide polymorphism data, confirming or rejecting paternity. Each plasma sample with a sufficient fetal cell-free DNA fraction was independently tested against the confirmed father and 1,820 random, unrelated males.
Results:
One of the 21 samples had insufficient fetal cell-free DNA. The test correctly confirmed paternity for the remaining 20 samples (100%) when tested against the biological father, with P values of <10−4. For the 36,400 tests using an unrelated male as the alleged father, 99.95% (36,382) correctly excluded paternity and 0.05% (18) were indeterminate. There were no miscalls.
Conclusion:
A noninvasive paternity test using informatics-based analysis of single-nucleotide polymorphism array measurements accurately determined paternity early in pregnancy.
Genet Med 2013:15(6):473–477
Similar content being viewed by others
Introduction
Reliable prenatal paternity testing requires direct chorionic villus sampling or amniotic fluid sampling via amniocentesis, both of which increase risk of harm to the mother and fetus. Amniocentesis carries a miscarriage risk of 1 in 300 to 1 in 500 (ref. 1) and a small risk of other complications.2,3 Chorionic villus sampling carries a similar miscarriage risk and a 1/3,000 risk of fetal limb reduction defects, especially when performed before 10 weeks of gestation.4,5 Although the American College of Obstetricians and Gynecologists recommends that these procedures be offered to all pregnant women,1 they are most often used when the mother is at a high risk for genetic defects, such as after a positive first-trimester screen.
The discovery of fetal cell-free DNA (cfDNA) in maternal blood suggested that a noninvasive paternity test could be developed, which would avoid the risks associated with invasive procedures.6 Although fetal cfDNA in maternal circulation was initially reported more than a decade ago,6,7,8 the development of a reliable, noninvasive paternity test using maternal peripheral blood has proved elusive because fetal cfDNA typically comprises <20% of the total cfDNA in maternal plasma and is highly fragmented.9 The limited amount of fetal cfDNA and its heavy dilution by maternal cfDNA present significant technical challenges. These can be overcome by coupling microarrays or high-throughput sequencing, which measure hundreds of thousands to millions of DNA segments, with sophisticated bioinformatics techniques to extract maximal information from the limited fetal genotype data obtained from maternal plasma cfDNA measurements.
We previously reported such a method, called Parental Support, which uses single-nucleotide polymorphism (SNP) microarray measurements from maternal and paternal DNA to improve the fidelity of noisy embryonic DNA genotype measurements.10 Although this method accurately identifies ploidy at all 24 chromosomes in single embryonic cells, it has not been adapted to paternity testing.
Here, we use a version of the Parental Support algorithm to determine whether the fetal components of the cfDNA SNP measurements made on cfDNA isolated from maternal plasma could be due to the alleged father. We report the results of testing this new method on 21 pregnant women, with each sample independently tested against the confirmed father as well as 1,820 unrelated individuals. We identify paternity with 100% accuracy as early as 6 weeks into pregnancy.
Materials and Methods
Inclusion criteria for enrolled women/couples were singleton pregnancies in the first or second trimester with known paternity. Couples were enrolled through in vitro fertilization clinics, obstetric offices, or online advertisements. All women who met the inclusion criteria were enrolled during the period from April to June 2011. Gestational ages ranged from 6 to 21 weeks, and paternity was confirmed in various ways ( Table 1 ). Nine of 21 samples had known parentage through control of fertilization during in vitro fertilization, with paternity reconfirmed through genotype-specific preimplantation genetic diagnosis. The remaining 12 samples had paternity confirmed by independent paternity testing on fetal or postnatal genetic material (DNA Diagnostic Center, Fairfield, OH). Written informed consent was obtained from all participants, and genetic samples were collected under an institutional review board–approved research protocol.
Maternal blood samples (~20 ml) were collected into cell-free blood tubes (Streck, Omaha, NE), and paternal blood samples (~5 ml) were collected into EDTA blood collection tubes. On average, 10 ml of plasma was isolated from each maternal sample via double centrifugation (1,600g for 10 min, tube transfer, centrifugation at 16,000g for 10 min). Maternal plasma cfDNA was isolated using the Qiagen (Valencia, CA) circulating nucleic acid kit and eluted in 45 µl buffer according to the manufacturer’s instructions. DNA termini were blunted in a reaction using 20 µl eluate in 1x NEB4 buffer, 0.42 mmol/l dNTP, and 2.5U T4 DNA polymerase (New England Biolabs, Ipswich, MA), incubated at 20 °C for 30 min, and the polymerase was denatured by incubating at 75 °C for 15 min. Three microliters of ligation mixture (0.5 µl 10x NEB 4, 1 µl 10 mmol/l adenosine triphosphate, 1 µl T4 polynucleotide kinase (New England Biolabs), 0.5 µl T4 DNA ligase (New England Biolabs)) was added, and samples were incubated at 16 °C for 24 h and then at 75 °C for 15 min. Test samples were transferred to a standard Illumina Infinium assay (Illumina, San Diego, CA), as were the maternal and alleged paternal genomic DNA samples. Briefly, 24 µl of DNA was whole-genome amplified at 37 °C for 20–24 h followed by fragmentation and precipitation. All subsequent procedures were performed in a Tecan EVO (Männedorf, Switzerland) unless otherwise specified. Precipitate was resuspended in microarray hybridization buffer, heat denatured, and transferred to Cyto12-SNP microarrays. The microarrays were incubated at 48 °C for at least 16 h, X-Stained (Infinium II Chemistry), washed, and scanned.
Microarray intensities were extracted using BeadStudio (Illumina) software. Fetal fraction in maternal plasma was determined by considering autosomal SNPs for which the mother is homozygous. The group of maternal homozygous SNPs was divided into subsets in which the fetus is either very likely or very unlikely to have the same genotype as the mother, on the basis of population frequencies. The fetal fraction is proportional to the difference of observed off-allele intensity between the two subsets.
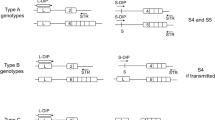
The genotyping results were analyzed using Parental Support.8 For each mother and alleged father combination, the method generated a test statistic that indicated how well the genotype of that possible father accounted for the fetal component of the genotypic measurements made on maternal plasma cfDNA. The test statistic was calculated separately for the mother with each of 5,000 unrelated males, creating an unrelated-male test statistic distribution for that case ( Figure 1 and Supplementary Figures S1–S19 online). The test statistic was then calculated for the mother and alleged father, and a single-hypothesis rejection test determined whether the alleged father’s test statistic fell under the distribution of test statistics created using the unrelated males. If the alleged father’s test statistic was excluded from the unrelated-male test statistic distribution, then the alleged father was determined to be the biological father. If the alleged father’s test statistic fell within the unrelated-male test statistic distribution, then it was determined that he was not the biological father.
The Parental Support method for paternity determination. Paternity test results for two separate women: (a) one with paternity inclusion and (b) one with paternity exclusion. The test statistic distribution is indicated in gray, the region indicating paternity inclusion (alleged fathers with P values <10−4) is indicated in green, the region indicating paternity exclusion (alleged fathers with P values >0.02) is indicated in beige, and the region indicating indeterminate paternity (alleged fathers with P values between 10−4 and 0.02) is indicated in black.
Specifically, a paternity inclusion (confirming paternity) was called when the P value of the alleged father’s test statistic according to the distribution of unrelated males was <10−4. An indeterminate result (no call) was called when the calculated P value fell between 10−4 and 0.02 ( Figure 1 ). A paternity exclusion was called when the calculated P value was >0.02. No determination was made when the fetal DNA fraction in maternal cfDNA was <2%. The set of unrelated individuals used to generate the expected distribution was comprised of individuals from a wide variety of racial backgrounds, and paternity determination was recalculated using different racial sets of unrelated individuals, including the race indicated for the alleged father. Inclusion and exclusion results were automatically generated by the informatics method; no human judgment was necessary.
Results
Twenty-one maternal blood samples with known paternity were tested using Parental Support ( Table 1 ). One sample could not be evaluated due to the low fraction of fetal cfDNA in the maternal plasma (<2%, Table 1 ). For each of the remaining 20 samples with sufficient fetal cfDNA, 1,821 independent paternity tests were run using the confirmed father and a random set of 1,820 unrelated male individuals, for a total of 36,420 paternity tests. We confirmed paternity in 100% (20/20) of the cases, with P values of <10−4 ( Table 1 ). The test accurately identified paternity as early as 6 weeks into pregnancy and with a low fraction of fetal cfDNA.
Of note, 99.95% (36,382/36,400) of the tests correctly excluded paternity using Parental Support. Only 18 (0.05%) were called indeterminate. None of the tested samples had an incorrect paternity determination.
Discussion
We report the first highly accurate, noninvasive, clinically available prenatal paternity test. Traditional postnatal paternity testing involves analysis of single tandem repeat sequences and comparison with alleged fathers. Because large amounts of intact genomic child DNA are available postnatally, measurements are robust, and only 15–20 single tandem repeat sequences are necessary to achieve highly accurate paternity results.11
Options are limited for determining paternity prenatally without amniocentesis or chorionic villus sampling, or very early in the preganancy. Although fetal cfDNA from maternal plasma is reliably measured,12 and various groups have reported the amplification and detection of fetal-specific alleles using polymerase chain reaction,13,14,15 these methods do not include sufficient fetal-specific alleles to measure paternity accurately and are not amenable to scaling of interrogated loci.
The use of cfDNA to correctly identify paternity was recently reported in 30 cases.16 However, that study only compared the known father to one unrelated male. If the actual father is not one of the two samples, this method may identify paternity in an individual who is simply more closely related than the other due to close genetic relationships in, for example, small ethnic subpopulations. This underscores the importance of testing against a large population of unrelated males; in this study, we tested against 5,000 random individuals.
Of greater concern is the fact that they do not report a per-test accuracy, instead reporting the chance of correctly guessing paternity in 30 cases, which is one in a billion—a number calculated by taking the chance of correctly guessing paternity per test (1/2) and raising it to the 30th power (30 tests). This combined statistic may be misleading and is not clinically useful, given that each test need not be highly accurate to provide this impressive number. For example, assuming 99% per-test accuracy (well below the 99.99% industry standard), the overall chance of guessing 30 tests correctly is still 74%.
By comparison, assuming a 99% per-test accuracy using Parental Support, the chance of correctly guessing all 36,402 calls is ~1 in 10158. Assuming 99.99% accuracy, the typical after-birth paternity testing accuracy level, the chance of correctly guessing all 36,402 calls is still only 2.6%. This suggests that the Parental Support method is >99.99% accurate.
It is important to note that a closely related male may return a false paternity inclusion. Preliminary results show that a paternal grandfather or uncle of the child in question returns test statistics that, although smaller than the actual father’s test statistic, have P values of <10−4 (data not shown); for this reason, samples from closely related alleged fathers must be measured for comparison to ensure correct paternity identification.
The method used herein obviates problems with traditional- and cfDNA-based methods by measuring autosomal SNPs using a high-throughput SNP array. A large amount of maternal cfDNA is present in the plasma measurements, which generally results in low signal-to-noise ratio when determining fetal genotype from cfDNA. Identifying the maternal genotype and fetal fraction allows us to predict the amount of maternal signal and adjust accordingly. Whereas each individual SNP measurement is noisy, aggregating data over 300,000 SNPs allows for highly accurate paternity determinations. This test provided accurate paternity confirmation as early as 6 weeks into pregnancy, with as low as 2.3% fetal cfDNA fraction, suggesting that paternity testing can be performed very early in pregnancy when fetal fractions are still very low. This is important when paternity is considered as a reason for termination of pregnancy.
This method is adaptable to other platforms, such as next-generation sequencing, which has recently been used to identify fetal cfDNA in maternal plasma.17 That study used massively parallel sequencing of maternal plasma cfDNA from a single subject, identifying the entire fetal and maternal genomes. Parental genotypes were included to assemble the fetal genome and determine maternal versus paternal SNP inheritance, and not in a statistical paternity test; the method lacks informatics-based analysis that contributes to the accuracy with which we identify and exclude paternity, and is more labor intensive, thereby not lending itself well to clinical applications. In addition, the maternal haplotype was inferred from chorionic villi sampling, thus negating the advantage of a noninvasive test. We are currently attempting to validate the efficacy of Parental Support using next-generation sequencing.
In summary, we report a highly accurate, noninvasive, clinically available prenatal paternity test. A larger study is warranted to accurately determine how early in pregnancy this method identifies paternity and to confirm efficacy in a larger cohort.
Disclosure
All authors are employees of Natera and/or hold stock or have options to hold stock in the company.
References
American College of Obstetricians and Gynecologists. Invasive prenatal testing for aneuploidy. ACOG Practice Bulletin No. 88, December 2007. Obstet Gynecol 2007;110:1459–1467.
Seeds JW . Diagnostic mid trimester amniocentesis: how safe? Am J Obstet Gynecol 2004;191:607–615.
Elchalal U, Shachar IB, Peleg D, Schenker JG . Maternal mortality following diagnostic 2nd-trimester amniocentesis. Fetal Diagn Ther 2004;19:195–198.
Brambati B, Simoni G, Travi M, et al. Genetic diagnosis by chorionic villus sampling before 8 gestational weeks: efficiency, reliability, and risks on 317 completed pregnancies. Prenat Diagn 1992;12:789–799.
Firth HV, Boyd PA, Chamberlain PF, MacKenzie IZ, Morriss-Kay GM, Huson SM . Analysis of limb reduction defects in babies exposed to chorionic villus sampling. Lancet 1994;343:1069–1071.
Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350:485–487.
Simpson JL, Elias S . Fetal cells in maternal blood: overview and historical perspective. Ann NY Acad Sci 1994;7:1–8.
Kazakov VI, Bozhkov VM, Linde VA, Repina MA, Mikhaı˘lov VM .Extracellular DNA in the blood of pregnant women. Tsitologia 1995;37:232–236.
Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011;342:c7401.
Johnson DS, Gemelos G, Baner J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod 2010;25:1066–1075.
Pena SD, Chakraborty R . Paternity testing in the DNA era. Trends Genet 1994;10:204–209.
Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM . Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999;64:218–224.
Zhu B, Sun QW, Lu YC, Sun MM, Wang LJ, Huang XH . Prenatal fetal sex diagnosis by detecting amelogenin gene in maternal plasma. Prenat Diagn 2005;25:577–581.
Wagner J, Dzijan S, Marjanovic D, Lauc G . Non-invasive prenatal paternity testing from maternal blood. Int J Legal Med 2009;123:75–79.
Lo YM, Bowell PJ, Selinger M, et al. Prenatal determination of fetal RhD status by analysis of peripheral blood of rhesus negative mothers. Lancet 1993;341:1147–1148.
Guo X, Bayliss P, Damewood M, et al. A noninvasive test to determine paternity in pregnancy. N Engl J Med 2012;366:1743–1745.
Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010;2:61ra91.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figure S1.
(TIFF 36498 kb)
Supplementary Figure S2.
(TIFF 36490 kb)
Supplementary Figure S3.
(TIFF 36490 kb)
Supplementary Figure S4.
(TIFF 36490 kb)
Supplementary Figure S5.
(TIFF 36489 kb)
Supplementary Figure S6.
(TIFF 36490 kb)
Supplementary Figure S7.
(TIFF 36490 kb)
Supplementary Figure S8.
(TIFF 36490 kb)
Supplementary Figure S9.
(TIFF 36490 kb)
Supplementary Figure S10.
(TIFF 36490 kb)
Supplementary Figure S11.
(TIFF 36490 kb)
Supplementary Figure S12.
(TIFF 36489 kb)
Supplementary Figure S13.
(TIFF 36489 kb)
Supplementary Figure S14.
(TIFF 36489 kb)
Supplementary Figure S15.
(TIFF 36490 kb)
Supplementary Figure S16.
(TIFF 36490 kb)
Supplementary Figure S17.
(TIFF 36490 kb)
Supplementary Figure S18.
(TIFF 36490 kb)
Supplementary Figure S19.
(TIFF 39796 kb)
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ryan, A., Baner, J., Demko, Z. et al. Informatics-based, highly accurate, noninvasive prenatal paternity testing. Genet Med 15, 473–477 (2013). https://doi.org/10.1038/gim.2012.155
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2012.155
Keywords
This article is cited by
-
Early noninvasive prenatal paternity testing by targeted fetal DNA analysis
Scientific Reports (2023)
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study
BMC Medical Genomics (2020)
-
Non-invasive prenatal paternity testing using a standard forensic genetic massively parallel sequencing assay for amplification of human identification SNPs
International Journal of Legal Medicine (2019)
-
Detection of short tandem repeats at 5 loci and amelogenin with cell-free fetal DNA as a specimen in the development of prenatal paternity diagnostic tests
Egyptian Journal of Forensic Sciences (2018)
-
An SNP panel for the analysis of paternally inherited alleles in maternal plasma using ion Torrent PGM
International Journal of Legal Medicine (2018)